| DPOGS200124 | ||
|---|---|---|
| Transcript | DPOGS200124-TA | 639 bp |
| Protein | DPOGS200124-PA | 212 aa |
| Genomic position | DPSCF300044 + 985518-986931 | |
| RNAseq coverage | 259x (Rank: top 41%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL015484 | 3e-55 | 48.10% | |
| Bombyx | BGIBMGA002401-TA | 1e-50 | 45.96% | |
| Drosophila | qm-PB | 1e-49 | 44.29% | |
| EBI UniRef50 | UniRef50_O95749 | 5e-46 | 46.67% | Geranylgeranyl pyrophosphate synthase n=73 Tax=Eukaryota RepID=GGPPS_HUMAN |
| NCBI RefSeq | XP_001605679.1 | 7e-50 | 47.37% | PREDICTED: similar to ENSANGP00000021639 [Nasonia vitripennis] |
| NCBI nr blastp | gi|156554759 | 1e-48 | 47.37% | PREDICTED: geranylgeranyl pyrophosphate synthase-like [Nasonia vitripennis] |
| NCBI nr blastx | gi|156554759 | 3e-47 | 47.55% | PREDICTED: geranylgeranyl pyrophosphate synthase-like [Nasonia vitripennis] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0008299 | 5.9e-25 | isoprenoid biosynthetic process | |
| KEGG pathway | nvi:100122074 | 2e-49 | ||
| K00804 (GGPS1) | maps-> | Terpenoid backbone biosynthesis | ||
| InterPro domain | [8-197] IPR017446 | 1.5e-60 | Polyprenyl synthetase-related | |
| [8-195] IPR008949 | 3.3e-41 | Terpenoid synthase | ||
| [19-194] IPR000092 | 5.9e-25 | Polyprenyl synthetase | ||
| Orthology group | ||||
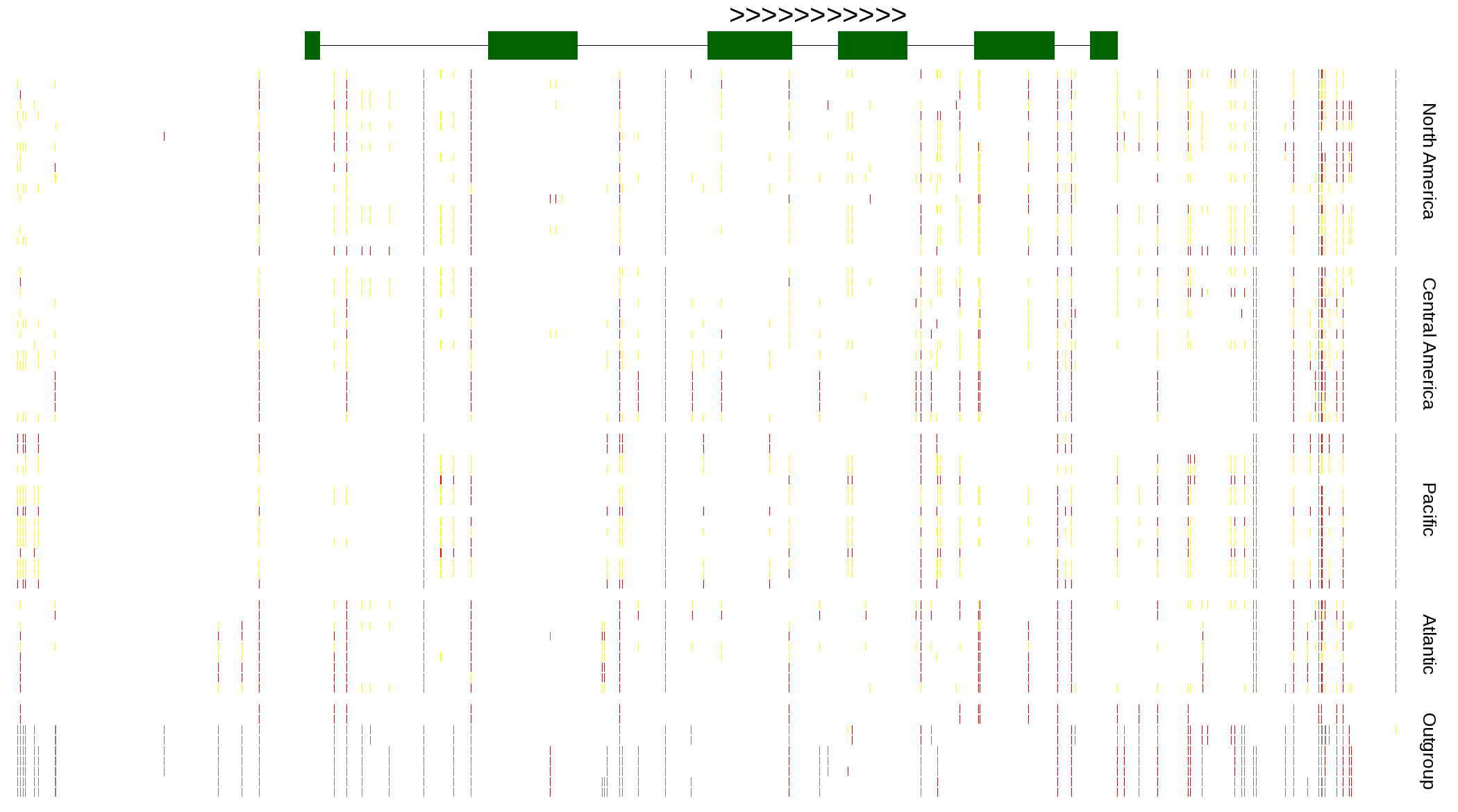
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS200124-TA
ATGGACTGTGAAAAAAATCTAGAAAAGGAGCTTCTTGCACCCTACATGCATTTGCTGCAAGTTAAAAGCAAGCAGTTTCGGAATAAAATCGTATTGGCCTTCAATTACTGGTTCAAGGTACCAGACGATCTAGTTCAAAATGCACTCAATGCGATGACAATGACATACAACGCAAGTTTATTGATCGATGATATTCAAGATAATTCAATAAAACGTCGAGGGATTGAAGCAGCTCATTGTATTTATGGAATTCCATTAACTATTAATACAGCAGTTCATGTTTTGGTTCTGGCCCTAAAACAAGAATTAGACCTCGTACCAGCCGCATTGGGTGGTAAAATATTTGTTGACCACGTTCTGGATGTAATTTACGGCCAAGGTGTTGAAATTTACTGGCGAGACAACTTTATGTGTCCGTCTGAAAAAGAGTATAAAGAAATGGTCCAGCAAAAAACTGGTGCCATGCTTTTATATGGAGTGAAATTAATACAAGTTTTAAGTGATGACAAAAGAAATTACGACGATTTTGTGAAACTTCTTGGTTTTTATTTCCAATTGAGAGACGACTACTGCAATTTAAAACAACTAGAAGCATTAGAGGAATACCCTGAAAAGGAAGATGAAAGTGTATGTGTCTGA
>DPOGS200124-PA
MDCEKNLEKELLAPYMHLLQVKSKQFRNKIVLAFNYWFKVPDDLVQNALNAMTMTYNASLLIDDIQDNSIKRRGIEAAHCIYGIPLTINTAVHVLVLALKQELDLVPAALGGKIFVDHVLDVIYGQGVEIYWRDNFMCPSEKEYKEMVQQKTGAMLLYGVKLIQVLSDDKRNYDDFVKLLGFYFQLRDDYCNLKQLEALEEYPEKEDESVCV-