| DPOGS200260 | ||
|---|---|---|
| Transcript | DPOGS200260-TA | 717 bp |
| Protein | DPOGS200260-PA | 238 aa |
| Genomic position | DPSCF300026 - 1124667-1125800 | |
| RNAseq coverage | 248x (Rank: top 42%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL005384 | 5e-113 | 87.85% | |
| Bombyx | % | |||
| Drosophila | mRpS7-PA | 6e-58 | 53.27% | |
| EBI UniRef50 | UniRef50_Q7Q1V7 | 5e-70 | 62.07% | AGAP009613-PA n=4 Tax=Endopterygota RepID=Q7Q1V7_ANOGA |
| NCBI RefSeq | XP_001649336.1 | 5e-72 | 59.91% | mitochondrial ribosomal protein S7 [Aedes aegypti] |
| NCBI nr blastp | gi|312381617 | 5e-74 | 65.67% | hypothetical protein AND_06035 [Anopheles darlingi] |
| NCBI nr blastx | gi|312381617 | 1e-72 | 65.67% | hypothetical protein AND_06035 [Anopheles darlingi] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0005840 | 1.4e-63 | ribosome | |
| GO:0006412 | 1.4e-63 | translation | ||
| GO:0005622 | 1.4e-63 | intracellular | ||
| GO:0003735 | 1.4e-63 | structural constituent of ribosome | ||
| KEGG pathway | cob:COB47_0737 | 5e-27 | ||
| K02992 (RP-S7, rpsG) | maps-> | Ribosome | ||
| InterPro domain | [65-238] IPR000235 | 1.4e-63 | Ribosomal protein S7 | |
| [78-237] IPR023798 | 2.9e-44 | Ribosomal protein S7 domain | ||
| Orthology group | MCL16101 | Multiple-copy universal gene | ||
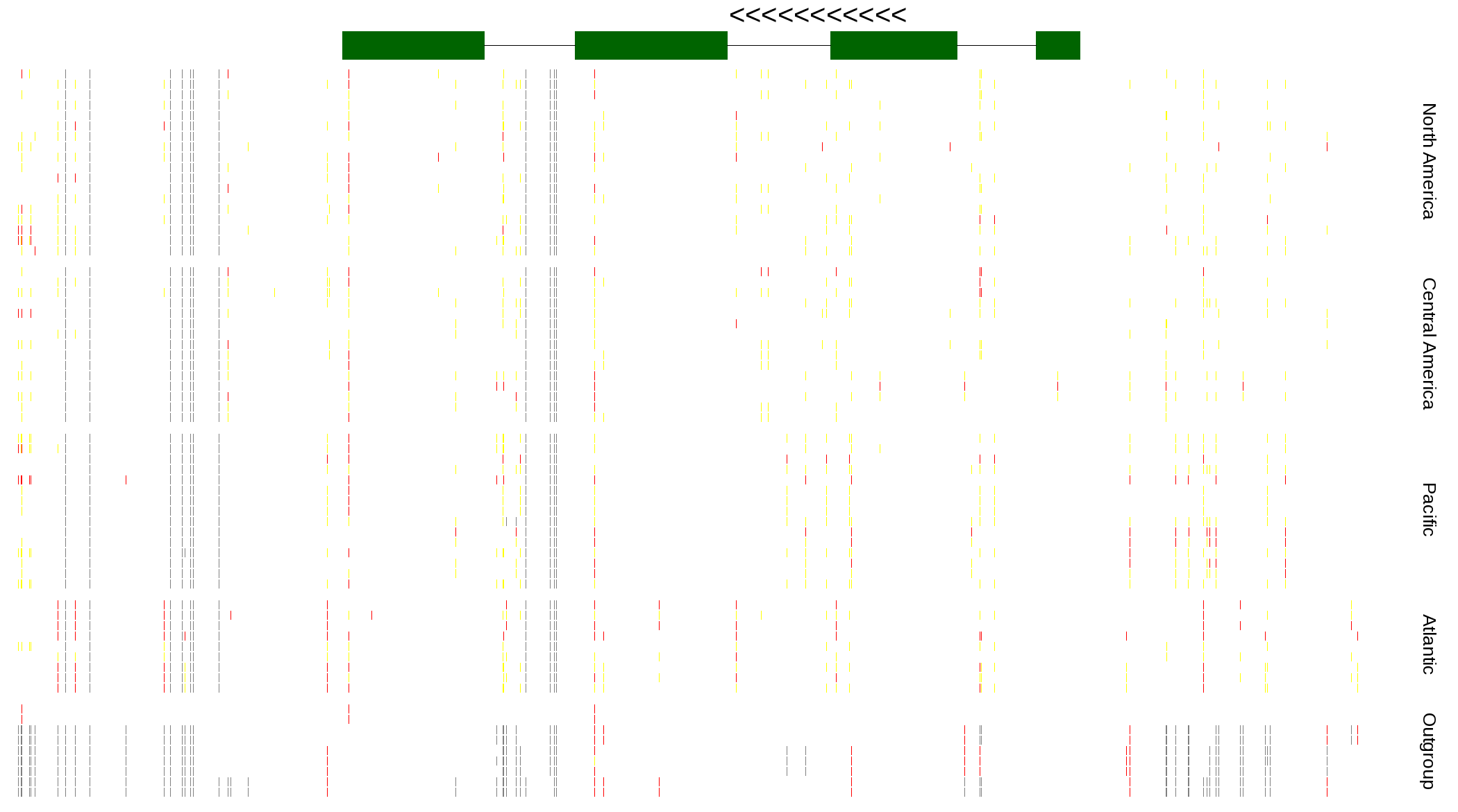
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS200260-TA
ATGAATAATATAAGGATAGCTTTAAGAGAAGTTATACCAAATAGACTTCGGACTACATATAGAAATCAATGGCCAAACTTAACGCAGTCCAGATTTTATCCCCGAGCCACGAGCTTTCCAGATTATTTTCAAAACCCAATATTTAGAAAAGAGGATCAAGCAAAATTGATAGAAAATAATGAATTGACTAAGCTGGCCGCGGTCCCTGTGAAACCACCTGCTGCTTATGAGACATCCTCAGTGTACCATGATGCACTTGTGGATAAAGTTATCAACCATGTAATGAAAATGGGCAACAAACAATTGGCTCGCAGTCTTGTTGAAAAAGCATTTGAGAACATCAAAAGGAAACAGATAGAGAGATATCATCTAGCTTCAACGCCTGAAGACAAGGCGAAAATTCTGTTGAACCCTAAAGATGTTTTACACAGAGCTGTTGAAAACTGTAAACCGCTGTTACAACTTTTACCAATTAAAAGAGGAGGTATCACATACCAGGTGCCAGGACCTATAACTGAGAAAAGGTCTCTATTTTTGGCTGTCAAATGGTTGTTAGATGCTTCAAATGACAAAGAAAGAACAGTACATTTTCCTGAACAATTTGCTTGGGAACTCCTTGATGCGGCTAACAATACAGGAAAGGTTGTGAAGAGAAAGCAGGACCTCCACCGTCTTTGTGAAGCCAACAGGGCTTATGCACATTATAGATGGCAATAA
>DPOGS200260-PA
MNNIRIALREVIPNRLRTTYRNQWPNLTQSRFYPRATSFPDYFQNPIFRKEDQAKLIENNELTKLAAVPVKPPAAYETSSVYHDALVDKVINHVMKMGNKQLARSLVEKAFENIKRKQIERYHLASTPEDKAKILLNPKDVLHRAVENCKPLLQLLPIKRGGITYQVPGPITEKRSLFLAVKWLLDASNDKERTVHFPEQFAWELLDAANNTGKVVKRKQDLHRLCEANRAYAHYRWQ-