| DPOGS200319 | ||
|---|---|---|
| Transcript | DPOGS200319-TA | 393 bp |
| Protein | DPOGS200319-PA | 130 aa |
| Genomic position | DPSCF300026 + 99800-100192 | |
| RNAseq coverage | 90x (Rank: top 63%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL003016 | 2e-51 | 70.40% | |
| Bombyx | BGIBMGA005615-TA | 6e-52 | 71.20% | |
| Drosophila | % | |||
| EBI UniRef50 | UniRef50_B1HYM6 | 4e-30 | 48.00% | Zinc-type alcohol dehydrogenase-like protein n=93 Tax=Bacteria RepID=B1HYM6_LYSSC |
| NCBI RefSeq | XP_001193453.1 | 9e-12 | 38.18% | PREDICTED: similar to auxin-induced protein [Strongylocentrotus purpuratus] |
| NCBI nr blastp | gi|332366218 | 3e-35 | 56.00% | alcohol dehydrogenase [Streptococcus sanguinis SK355] |
| NCBI nr blastx | gi|332366218 | 7e-34 | 56.00% | alcohol dehydrogenase [Streptococcus sanguinis SK355] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0008270 | 5.1e-33 | zinc ion binding | |
| GO:0055114 | 5.1e-33 | oxidation-reduction process | ||
| GO:0016491 | 5.1e-33 | oxidoreductase activity | ||
| KEGG pathway | reh:H16_B1699 | 8e-32 | ||
| K00001 (E1.1.1.1, adh) | maps-> | Drug metabolism - cytochrome P450 | ||
| Glycolysis / Gluconeogenesis | ||||
| Fatty acid metabolism | ||||
| 3-Chloroacrylic acid degradation | ||||
| Tyrosine metabolism | ||||
| Metabolism of xenobiotics by cytochrome P450 | ||||
| 1- and 2-Methylnaphthalene degradation | ||||
| Retinol metabolism | ||||
| InterPro domain | [1-125] IPR002085 | 5.1e-33 | Alcohol dehydrogenase superfamily, zinc-type | |
| [1-130] IPR011032 | 4.8e-25 | GroES-like | ||
| [31-89] IPR013154 | 2.1e-08 | Alcohol dehydrogenase GroES-like | ||
| Orthology group | MCL21138 | Lepidoptera specific | ||
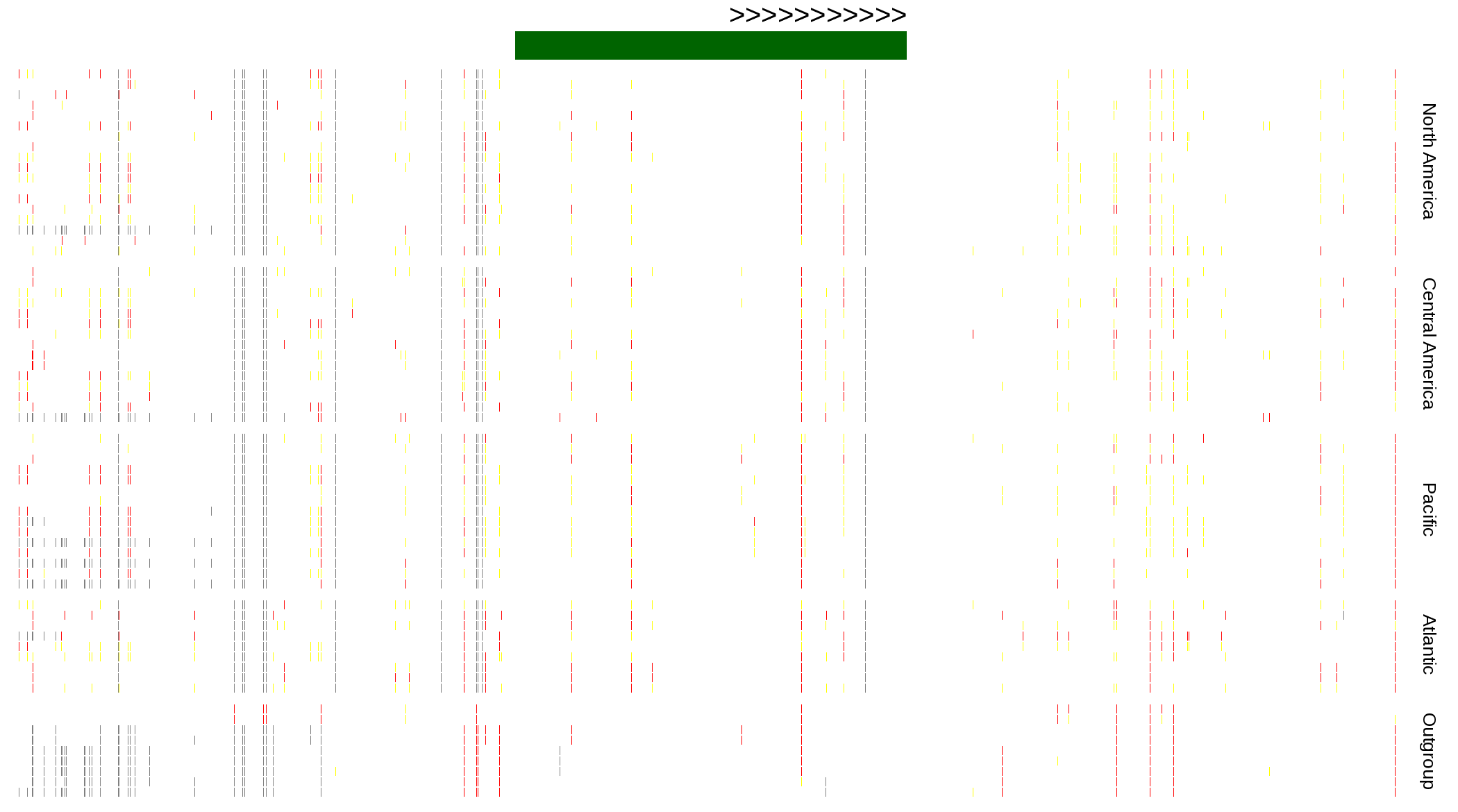
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS200319-TA
ATGAAAGCCGTAGGCTTATATAAATATCTGCCTCTGACTGACCCAAACTGTTTGTTGGATTTAGAATTAAATCTTCCCATGATTAAAAAAGACGAGGTTTTGGTGGAAGTGAAAGCAATCTCGGTGAATCCTATTGACACAAAACTACGATCCCCGAAACCCATTATTGAAACTAGCCCTCGAATATTAGGTTGGGATGGTTCAGGCACTGTTGTTGGAAGAGGTGACGGTTGTAAAATATTTAATATTGGTGACCCTGTATTTTTCACTGGAGATTTAAGGAAAAACGGTTCCAATGCACAATATGTTGCACTTAATGAGATCATGGTAGGTTCAAAACCCAAAAATCTGACATTTGAAGAAGCAGCAGCCATGATTGTTAATCATCAATAG
>DPOGS200319-PA
MKAVGLYKYLPLTDPNCLLDLELNLPMIKKDEVLVEVKAISVNPIDTKLRSPKPIIETSPRILGWDGSGTVVGRGDGCKIFNIGDPVFFTGDLRKNGSNAQYVALNEIMVGSKPKNLTFEEAAAMIVNHQ-