| DPOGS201046 | ||
|---|---|---|
| Transcript | DPOGS201046-TA | 465 bp |
| Protein | DPOGS201046-PA | 154 aa |
| Genomic position | DPSCF300299 + 126703-128011 | |
| RNAseq coverage | 83x (Rank: top 64%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL005362 | 1e-59 | 61.86% | |
| Bombyx | BGIBMGA012485-TA | 3e-51 | 54.64% | |
| Drosophila | CG11899-PA | 1e-36 | 44.62% | |
| EBI UniRef50 | UniRef50_UPI0000E49F4C | 3e-34 | 42.11% | UPI0000E49F4C related cluster n=1 Tax=unknown RepID=UPI0000E49F4C |
| NCBI RefSeq | NP_001040269.1 | 6e-52 | 56.70% | phosphoserine aminotransferase 1 [Bombyx mori] |
| NCBI nr blastp | gi|309401864 | 2e-54 | 60.00% | phosphoserine aminotransferase [Antheraea pernyi] |
| NCBI nr blastx | gi|309401864 | 7e-52 | 60.00% | phosphoserine aminotransferase [Antheraea pernyi] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0003824 | 2.1e-36 | catalytic activity | |
| GO:0030170 | 2.1e-36 | pyridoxal phosphate binding | ||
| GO:0008152 | 1.3e-09 | metabolic process | ||
| KEGG pathway | cqu:CpipJ_CPIJ008287 | 6e-38 | ||
| K00831 (E2.6.1.52, serC) | maps-> | Vitamin B6 metabolism | ||
| Glycine, serine and threonine metabolism | ||||
| InterPro domain | [47-145] IPR015421 | 2.1e-36 | Pyridoxal phosphate-dependent transferase, major region, subdomain 1 | |
| [9-144] IPR015424 | 1.7e-26 | Pyridoxal phosphate-dependent transferase, major domain | ||
| [61-111] IPR000192 | 1.3e-09 | Aminotransferase, class V/Cysteine desulfurase | ||
| Orthology group | ||||
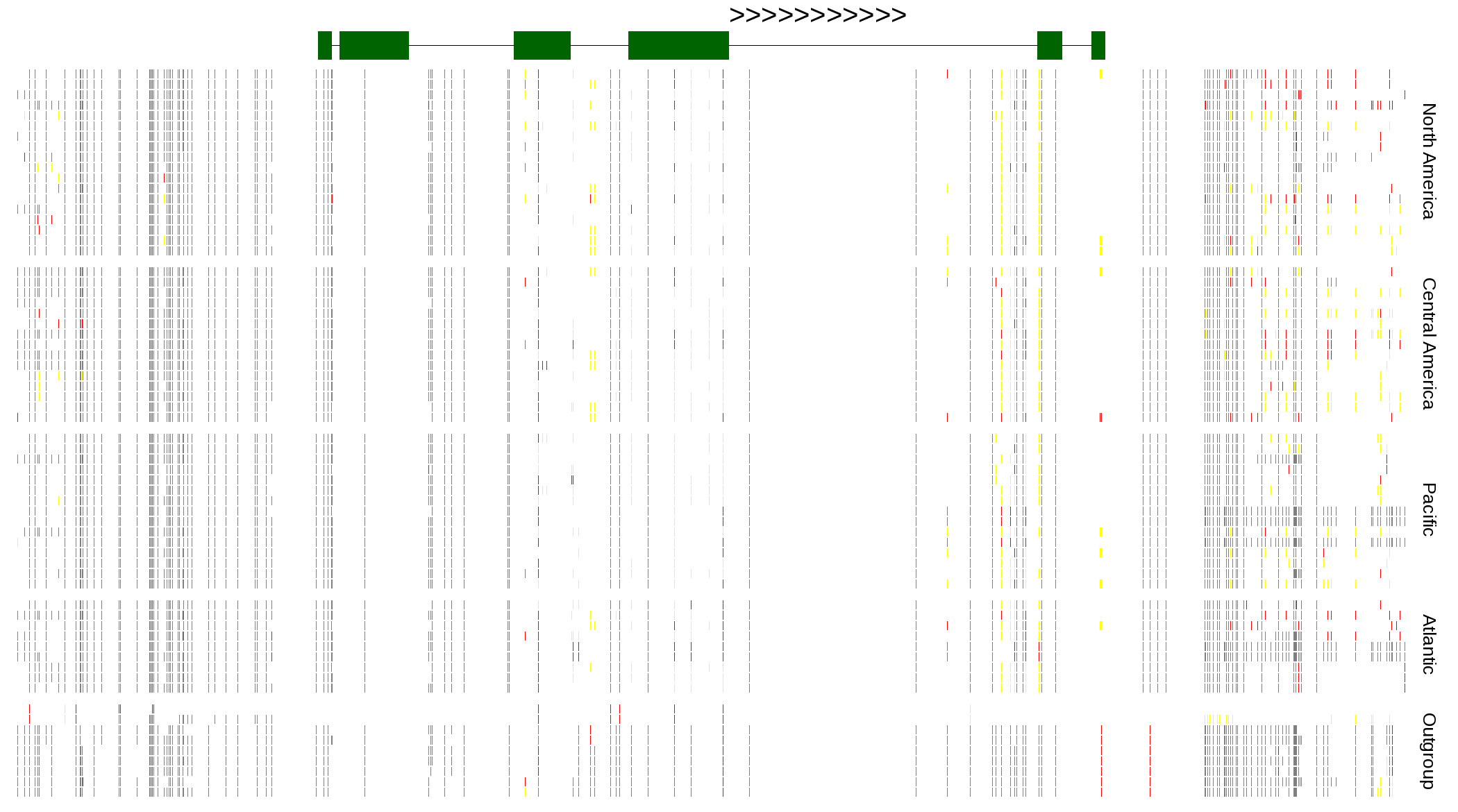
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS201046-TA
ATGGAACTGCAAATTGACAAAACAAATGTCCCGAACAACTACAAAATACTATTTCTATCTGGCGGCGGTCAGGGACAATTTGCTGCGGTGCCACTAAATTTAATATCACGTACTGGTTCAGCCGACTATGTAGTGACTGGAATAGAATTTGATTTTATCCCGGACACTAAAGTTGTCCCACTGGTTGCTGATATGTCTTCCAATTTTATGTCCAAGAAAATTGATGTTTCTAAATTTGGAGTCATTTACGGCGGTGCTCAAAAGAACATAGGTACTTCTGGTGTTGCTCTGGTTATAGTGAGGGAAGATCTTTTAAACCAGGCGCTTCCCATTTGCCCCTCAATATTGGATTGGACCATCAATGCGAAAGCGGATTCCATTCCTGACACACCTCCGATGTTTGTTATGGGACGACTGTTCCAATGGATCGACAGGCAAGACAATTGCCAAGAAAGACAGAAGTAA
>DPOGS201046-PA
MELQIDKTNVPNNYKILFLSGGGQGQFAAVPLNLISRTGSADYVVTGIEFDFIPDTKVVPLVADMSSNFMSKKIDVSKFGVIYGGAQKNIGTSGVALVIVREDLLNQALPICPSILDWTINAKADSIPDTPPMFVMGRLFQWIDRQDNCQERQK-