| DPOGS201197 | ||
|---|---|---|
| Transcript | DPOGS201197-TA | 669 bp |
| Protein | DPOGS201197-PA | 222 aa |
| Genomic position | DPSCF300262 + 224554-229724 | |
| RNAseq coverage | 3995x (Rank: top 3%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL004742 | 3e-62 | 66.48% | |
| Bombyx | BGIBMGA014241-TA | 6e-92 | 81.91% | |
| Drosophila | CG34325-PA | 3e-48 | 49.43% | |
| EBI UniRef50 | UniRef50_Q9U6W9 | 8e-88 | 81.42% | Death-associated LIM only protein DALP n=5 Tax=Endopterygota RepID=Q9U6W9_MANSE |
| NCBI RefSeq | XP_001965740.1 | 3e-49 | 54.02% | GF22290 [Drosophila ananassae] |
| NCBI nr blastp | gi|312451917 | 4e-90 | 87.15% | death-associated LIM-only protein [Helicoverpa armigera] |
| NCBI nr blastx | gi|312451917 | 2e-98 | 87.15% | death-associated LIM-only protein [Helicoverpa armigera] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0008270 | 2.5e-19 | zinc ion binding | |
| KEGG pathway | api:100162638 | 2e-41 | ||
| K05760 (PXN) | maps-> | Chemokine signaling pathway | ||
| Regulation of actin cytoskeleton | ||||
| Leukocyte transendothelial migration | ||||
| Bacterial invasion of epithelial cells | ||||
| Focal adhesion | ||||
| VEGF signaling pathway | ||||
| InterPro domain | [112-173] IPR001781 | 2.5e-19 | Zinc finger, LIM-type | |
| Orthology group | MCL17891 | Insect specific | ||
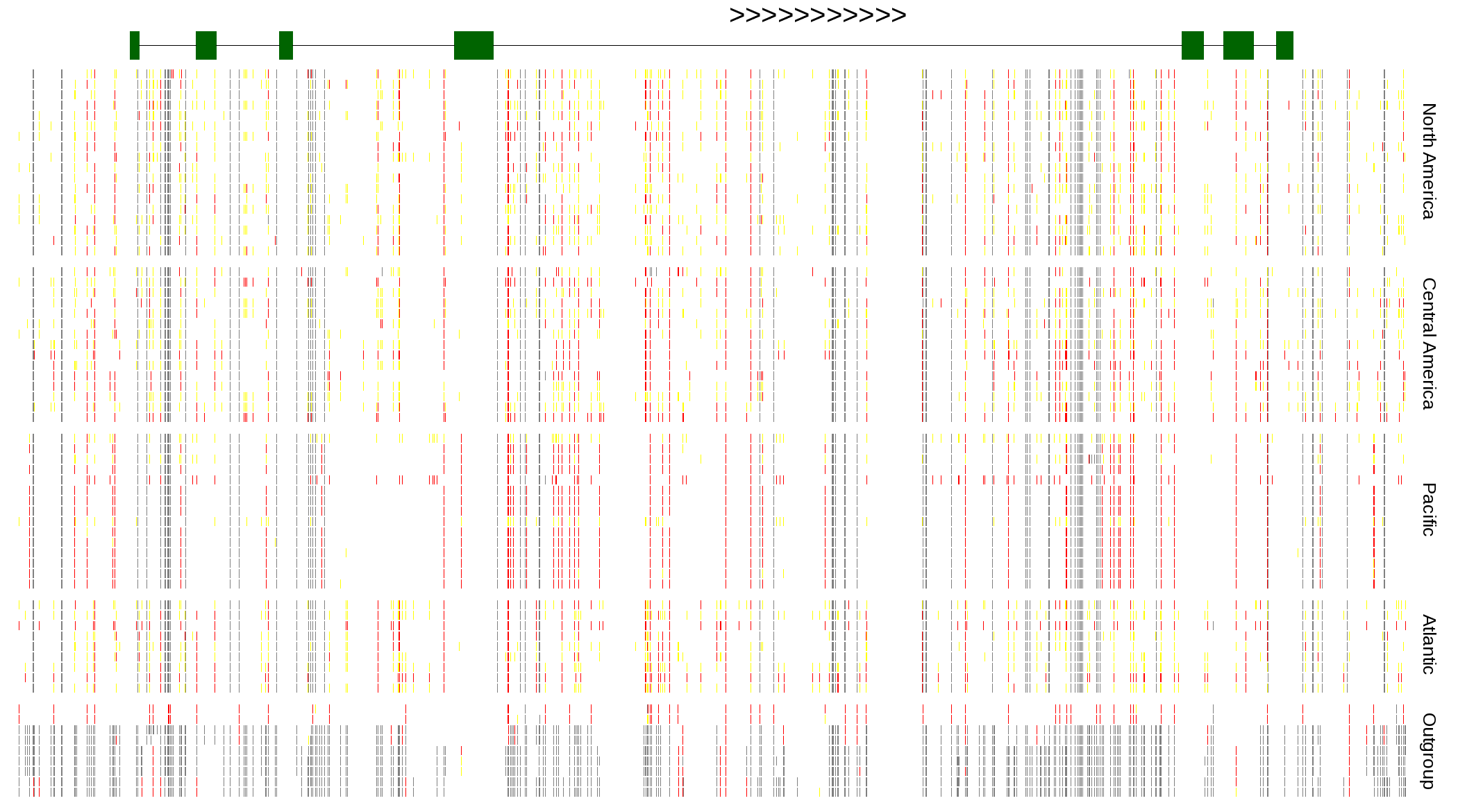
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS201197-TA
ATGTTACAACTAATTAATAATATTGAAAGTAAAACGACTAGATGCCAAACGTCTATTGTCACCGGTAACGCTATCGGTGCTATCAGTATACAATATTTATCAATCACCGAAGTGATGTCACATTTGAGGTCAACCAAAGACGGAGGGCCGGTCGTTTGTAACAGCTGTAATGGTGCTATCCAGGGCAGAATAGTGACAGCCCTGAACAAGAAGTGGCACCCAGAGCACTTCACCTGCAACACCTGTCACAAGCCTATAGACGGAGCCAAGTTCCACCAGCACGACGGAGGTATACACTGCGTCATCTGCTACGCTAAATATCACAGCCCGAGGTGCCATGGGTGTGGGGATCCCATCACTGATCGTGTGATCCAAGCTCTGGGTGTGTCCTGGCACGCTCACCACTTCGTGTGTGGTGGCTGTAAGAAGGAGCTCGGAGGAGGTGGCTTTATGGAACAGGCCGGTCGTCCATACTGCTCGTCCTGCTACGCGGACAAGTTCGCAGCGAGATGCGCCGGCTGCGCGTCCCCTATTGTGGACAAAGCGATCATCGCGCTCGACAACAAGTGGCACCGCGACTGCTTCACTTGCACCAAATGCCGGAACCCAGTAACCGACTCAACGTTCTCTGTGCTGGACAACAAGCCGCTGTGCGGTAAATGCGCTTGA
>DPOGS201197-PA
MLQLINNIESKTTRCQTSIVTGNAIGAISIQYLSITEVMSHLRSTKDGGPVVCNSCNGAIQGRIVTALNKKWHPEHFTCNTCHKPIDGAKFHQHDGGIHCVICYAKYHSPRCHGCGDPITDRVIQALGVSWHAHHFVCGGCKKELGGGGFMEQAGRPYCSSCYADKFAARCAGCASPIVDKAIIALDNKWHRDCFTCTKCRNPVTDSTFSVLDNKPLCGKCA-