| DPOGS201291 | ||
|---|---|---|
| Transcript | DPOGS201291-TA | 534 bp |
| Protein | DPOGS201291-PA | 177 aa |
| Genomic position | DPSCF300176 - 460487-461966 | |
| RNAseq coverage | 582x (Rank: top 22%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL012410 | 3e-99 | 94.35% | |
| Bombyx | BGIBMGA003040-TA | 1e-83 | 88.61% | |
| Drosophila | % | |||
| EBI UniRef50 | UniRef50_UPI0002246587 | 6e-26 | 36.09% | UPI0002246587 related cluster n=1 Tax=unknown RepID=UPI0002246587 |
| NCBI RefSeq | XP_001608134.1 | 1e-26 | 36.09% | PREDICTED: similar to ENSANGP00000028549 [Nasonia vitripennis] |
| NCBI nr blastp | gi|328711820 | 7e-27 | 44.37% | PREDICTED: hypothetical protein LOC100569183 [Acyrthosiphon pisum] |
| NCBI nr blastx | gi|328711820 | 3e-26 | 44.37% | PREDICTED: hypothetical protein LOC100569183 [Acyrthosiphon pisum] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0005515 | 1e-12 | protein binding | |
| GO:0003677 | 3.3e-11 | DNA binding | ||
| KEGG pathway | ||||
| InterPro domain | [20-82] IPR009057 | 1e-12 | Homeodomain-like | |
| [27-62] IPR007889 | 3.3e-11 | Helix-turn-helix, Psq | ||
| [87-150] IPR006600 | 1.6e-06 | Pogo transposase / Cenp-B / PDC2, DNA-binding HTH domain | ||
| Orthology group | MCL20678 | Insect specific | ||
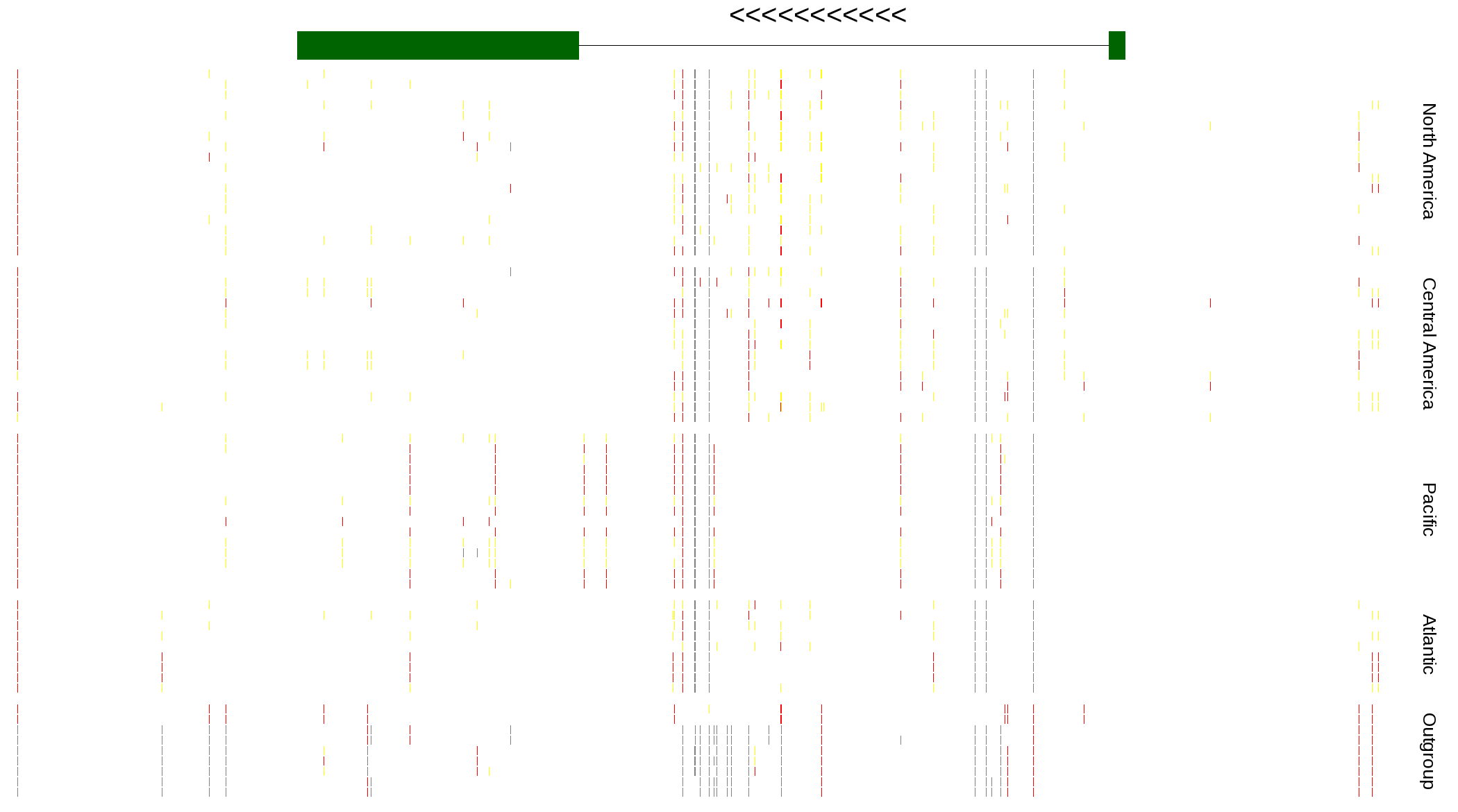
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS201291-TA
ATGCGGTGTGCGGTCGCCAACACGGTTACGACAGTCATGGTAAGGAAATACGAAAGAATGAGCGGACGCCAATCCTGGAGTGAAGAAGAAATGGCCAAAGCTGTGGCAGCCGTGGTGTCTGGCAAAATGGGCTACAAACTGGCAGCACGGACGTACCACATACCACGATCAACACTCCAGAGAAGAGCGAGCAAAGTACGATACCAGCAACCAGACGATCCCAAACCCATGATGGGGCATTATAGACGTGTGTTCACTGAGAGTCAAGAGAAAGACTTGGTTGGCTATATAAAGAGCATGGAGAAATATTTCATGGGAGTATCCCGGAGGGATATTAGGGAGCTGGCATTCCAATACGCCGAAGACAACAATCTAAACCATCCCTTCGACGCTAACACTCGGATGGCCGGCGAGGATTGGGTGAGGAACTTCTTAAAGCGGAATCCAGAACTGCTCCACAAATCAGAAAAAGACTACGAGTTGGAACCGGTGAACTTCGATCAGTTCTACCACTTCATGTGTCAATCTTCATGA
>DPOGS201291-PA
MRCAVANTVTTVMVRKYERMSGRQSWSEEEMAKAVAAVVSGKMGYKLAARTYHIPRSTLQRRASKVRYQQPDDPKPMMGHYRRVFTESQEKDLVGYIKSMEKYFMGVSRRDIRELAFQYAEDNNLNHPFDANTRMAGEDWVRNFLKRNPELLHKSEKDYELEPVNFDQFYHFMCQSS-