| DPOGS201420 | ||
|---|---|---|
| Transcript | DPOGS201420-TA | 525 bp |
| Protein | DPOGS201420-PA | 174 aa |
| Genomic position | DPSCF300006 - 1640900-1642195 | |
| RNAseq coverage | 82x (Rank: top 64%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL009070 | 3e-67 | 67.24% | |
| Bombyx | BGIBMGA002716-TA | 1e-64 | 65.12% | |
| Drosophila | CG5009-PA | 7e-32 | 41.86% | |
| EBI UniRef50 | UniRef50_D2SNX9 | 5e-52 | 58.33% | Acyl-CoA oxidase (Fragment) n=2 Tax=Heliothis virescens RepID=D2SNX9_HELVI |
| NCBI RefSeq | XP_973660.1 | 9e-47 | 54.07% | PREDICTED: similar to acyl-CoA oxidase [Tribolium castaneum] |
| NCBI nr blastp | gi|260907935 | 3e-55 | 63.92% | acyl-CoA oxidase [Heliothis virescens] |
| NCBI nr blastx | gi|260907935 | 6e-52 | 63.92% | acyl-CoA oxidase [Heliothis virescens] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0006635 | 1.9e-48 | fatty acid beta-oxidation | |
| GO:0003997 | 1.9e-48 | acyl-CoA oxidase activity | ||
| GO:0005777 | 1.9e-48 | peroxisome | ||
| GO:0055114 | 1.9e-48 | oxidation-reduction process | ||
| GO:0016627 | 8.1e-45 | oxidoreductase activity, acting on the CH-CH group of donors | ||
| KEGG pathway | tca:662475 | 2e-46 | ||
| K00232 (E1.3.3.6, ACOX1, ACOX3) | maps-> | Peroxisome | ||
| Fatty acid metabolism | ||||
| Biosynthesis of unsaturated fatty acids | ||||
| PPAR signaling pathway | ||||
| alpha-Linolenic acid metabolism | ||||
| InterPro domain | [2-171] IPR002655 | 1.9e-48 | Acyl-CoA oxidase, C-terminal | |
| [3-174] IPR009075 | 8.1e-45 | Acyl-CoA dehydrogenase/oxidase C-terminal | ||
| [85-139] IPR023570 | 2.5e-12 | Acyl-CoA oxidase, peroxisomal | ||
| Orthology group | ||||
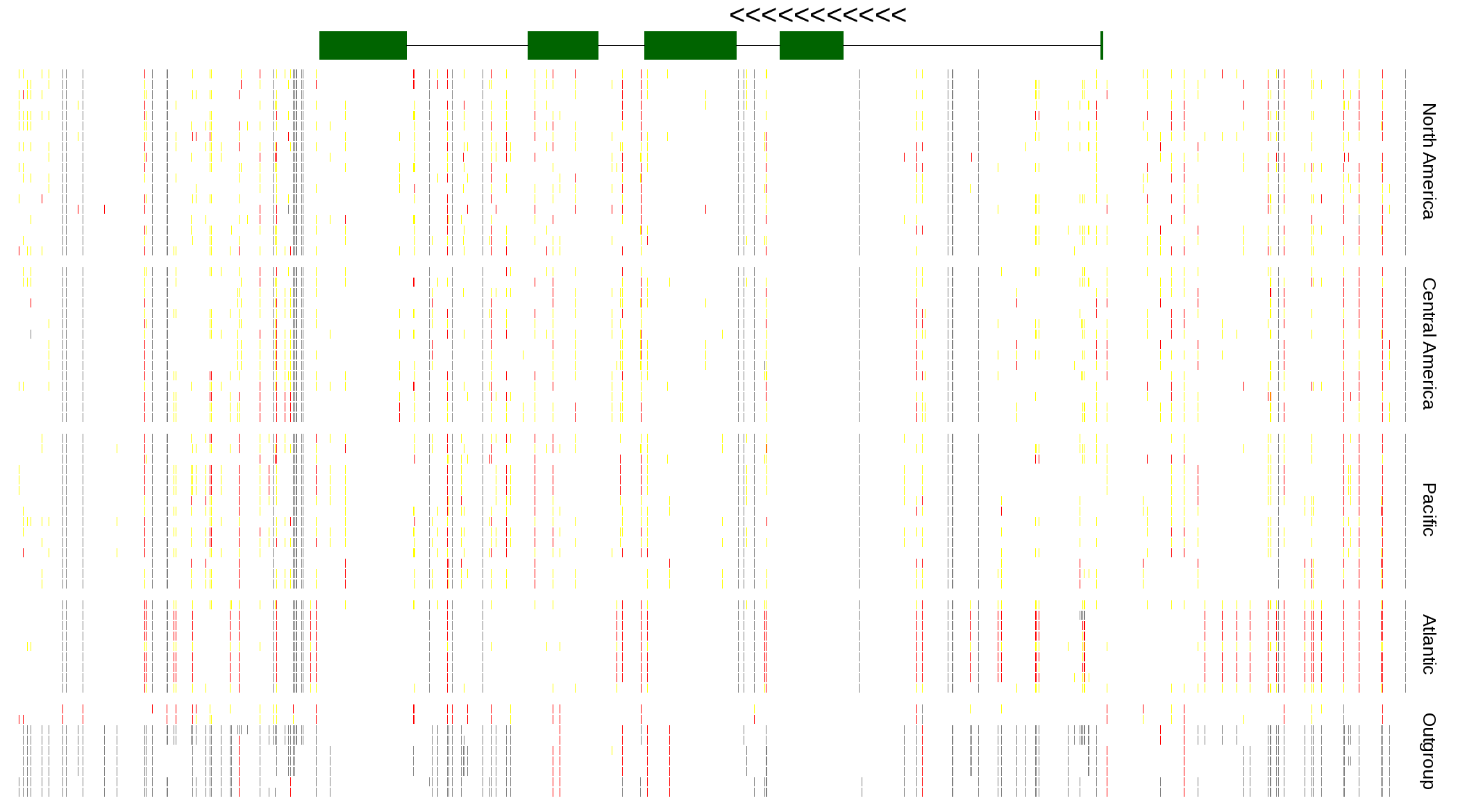
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS201420-TA
ATGAAAAAAATTCAATCTTGCGTAAAGGATATGGAAAATCGGGTTTCATCTGGAACATCATTTGAGGATGCTTGGAATATGAGTTCAGTACAATTGGTTTCAGCAGCAGAGGCGCATTGTAGAGTTATAATTCTGTCAACATTCAACGAGGAAATAAAAGAGAAAAGTAAAAGTGCGTCACCAGAACTAGCCGTCGTACTTAACCAGCTATTAGAATTATACACAGTCTATTGGGCACTTGAGAAATCTGGTGATTTATTGCAGTATACATGTATAAGTAAGGAGGACTTGGAGGGATTGCGTTGGTGGTACGAAGATATTCTTATACAGCTTCGACCCAACGCCGTTGGTGTAGTTGACGCTTTTGATATTAGAGACGATTTTCTGCATTCAACACTGGGAGCTTATGATGGTCGTGTCTACGAAAGGTTGATGGAAGAAGCACTCAAATGTCCTCTAAATGCTGATGTCGTAAATGTCTCTTTTCACAAATATCTGAAACCATTCTTGCGAGGGAAACTGTGA
>DPOGS201420-PA
MKKIQSCVKDMENRVSSGTSFEDAWNMSSVQLVSAAEAHCRVIILSTFNEEIKEKSKSASPELAVVLNQLLELYTVYWALEKSGDLLQYTCISKEDLEGLRWWYEDILIQLRPNAVGVVDAFDIRDDFLHSTLGAYDGRVYERLMEEALKCPLNADVVNVSFHKYLKPFLRGKL-