| DPOGS201525 | ||
|---|---|---|
| Transcript | DPOGS201525-TA | 585 bp |
| Protein | DPOGS201525-PA | 194 aa |
| Genomic position | DPSCF300006 + 1269230-1277321 | |
| RNAseq coverage | 230x (Rank: top 44%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL009093 | 4e-58 | 53.57% | |
| Bombyx | BGIBMGA002704-TA | 8e-67 | 64.24% | |
| Drosophila | NLaz-PA | 2e-33 | 40.45% | |
| EBI UniRef50 | UniRef50_E2ABN0 | 6e-53 | 50.27% | Apolipoprotein D n=14 Tax=Endopterygota RepID=E2ABN0_CAMFO |
| NCBI RefSeq | XP_001607285.1 | 6e-55 | 57.40% | PREDICTED: similar to ENSANGP00000012317 [Nasonia vitripennis] |
| NCBI nr blastp | gi|383848263 | 4e-55 | 52.57% | PREDICTED: apolipoprotein D-like [Megachile rotundata] |
| NCBI nr blastx | gi|383848263 | 1e-54 | 53.18% | PREDICTED: apolipoprotein D-like [Megachile rotundata] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0005488 | 4.9e-58 | binding | |
| GO:0008289 | 2e-10 | lipid binding | ||
| GO:0006810 | 2e-10 | transport | ||
| GO:0005215 | 2e-10 | transporter activity | ||
| KEGG pathway | ||||
| InterPro domain | [17-191] IPR012674 | 4.9e-58 | Calycin | |
| [25-190] IPR011038 | 1.6e-54 | Calycin-like | ||
| [7-195] IPR022271 | 2.6e-51 | Lipocalin, ApoD type | ||
| [46-189] IPR000566 | 1.9e-20 | Lipocalin/cytosolic fatty-acid binding protein domain | ||
| [22-36] IPR002969 | 2e-10 | Apolipoprotein D | ||
| Orthology group | MCL16218 | Patchy | ||
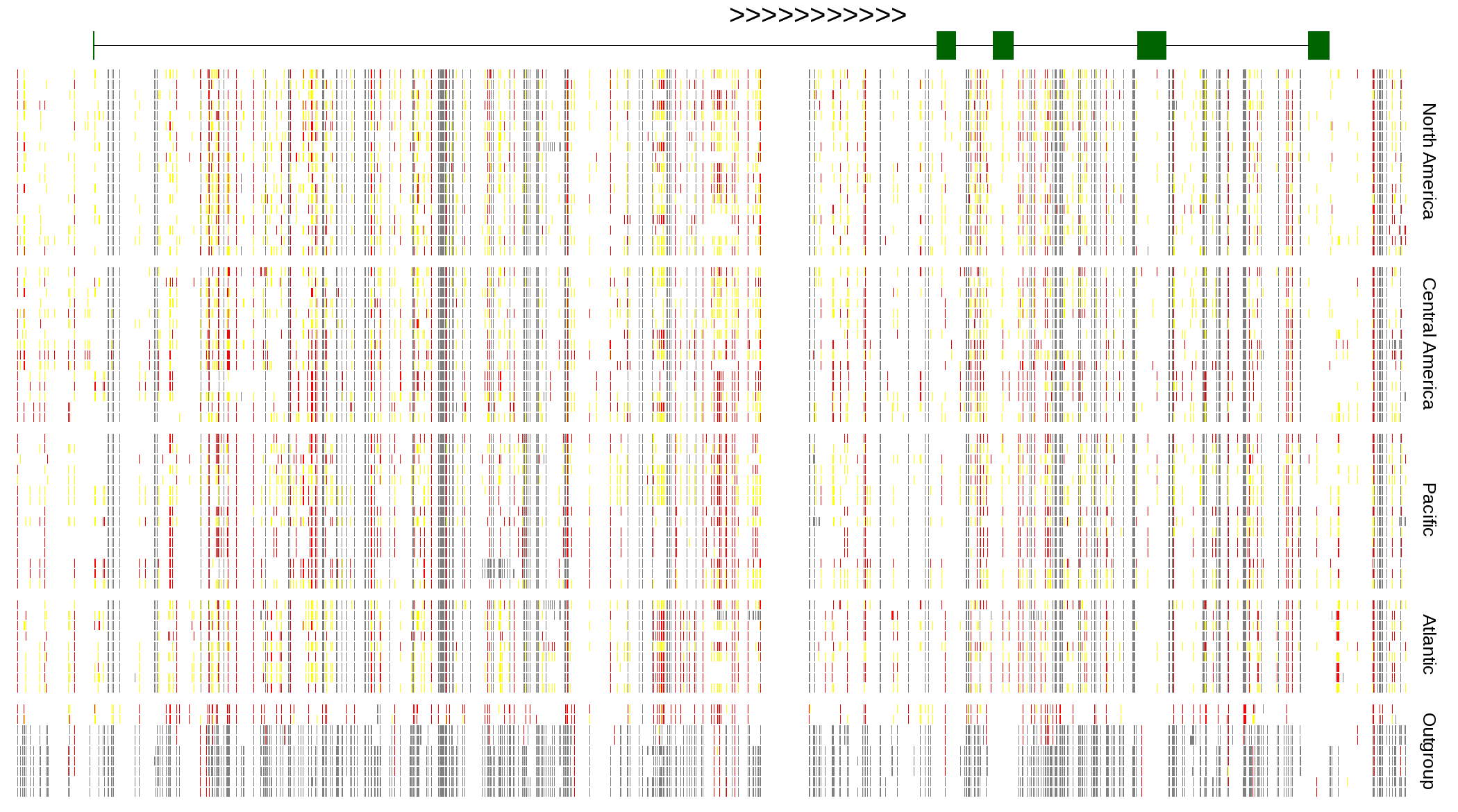
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS201525-TA
ATGATGTCCGTGATCCGCGCGATAGCTGTGTTCTTGTGCGGGTTGTCTTGGACAGTTTGCAAGGCACAGATTTTGTTCCCAGGAGCATGCCCTGATGTGGCCGCGATGAGCGACTTTGATCCCAGCCGTTATTTGGGGAAATGGTACGAAGCTGAAAAATACTTCGCAGCATTTGAGCTCGGTGGTGCCTGCATCACCGCCAATTACAAACTAAAGGATAATGGCGCCATCAGTGTTGTCAACGAGCAGTTCAGCTTATTAACCGGCACGAAAAAGTCTATAACCGGTGAAGCTGTTCAAGTCAGTCGCTCGGAGCCAGCCAAGCTATCAGTGACCTTCTCTTCTCTGCCGGTCAACATACCGGCCCCCTACTGGATCGTGAGCACAGACTATGACTCCTACGCTCTAATATGGAGCTGCTACGACTTTGGGATATTTCATACAAGAAATGCTTGGATCCTGACTAGACAAAGAAAGCCTTCAGCATCGGTTTTGGACGAGGCCTACTCGGCCGCTGACAAGAACAACATCAACAGGTCCTACTTCATGAGGACGGATCAAACAAACTGCTCCGAGGATTCTTAA
>DPOGS201525-PA
MMSVIRAIAVFLCGLSWTVCKAQILFPGACPDVAAMSDFDPSRYLGKWYEAEKYFAAFELGGACITANYKLKDNGAISVVNEQFSLLTGTKKSITGEAVQVSRSEPAKLSVTFSSLPVNIPAPYWIVSTDYDSYALIWSCYDFGIFHTRNAWILTRQRKPSASVLDEAYSAADKNNINRSYFMRTDQTNCSEDS-