| DPOGS201627 | ||
|---|---|---|
| Transcript | DPOGS201627-TA | 786 bp |
| Protein | DPOGS201627-PA | 261 aa |
| Genomic position | DPSCF300525 + 53831-54616 | |
| RNAseq coverage | 4x (Rank: top 88%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL003139 | 4e-60 | 46.80% | |
| Bombyx | BGIBMGA010193-TA | 6e-51 | 41.11% | |
| Drosophila | CG31548-PA | 9e-49 | 42.63% | |
| EBI UniRef50 | UniRef50_D2SNW1 | 1e-48 | 42.86% | Hydroxybutyrate dehydrogenase n=7 Tax=Obtectomera RepID=D2SNW1_HELVI |
| NCBI RefSeq | XP_001955231.1 | 1e-47 | 43.43% | GF18656 [Drosophila ananassae] |
| NCBI nr blastp | gi|260908006 | 5e-48 | 42.86% | hydroxybutyrate dehydrogenase [Heliothis virescens] |
| NCBI nr blastx | gi|260908006 | 1e-48 | 42.40% | hydroxybutyrate dehydrogenase [Heliothis virescens] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0005488 | 1.4e-73 | binding | |
| GO:0008152 | 6.4e-23 | metabolic process | ||
| GO:0016491 | 6.4e-23 | oxidoreductase activity | ||
| KEGG pathway | cpo:COPRO5265_1377 | 4e-33 | ||
| K00059 (fabG) | maps-> | Biosynthesis of unsaturated fatty acids | ||
| Fatty acid biosynthesis | ||||
| InterPro domain | [1-254] IPR016040 | 1.4e-73 | NAD(P)-binding domain | |
| [7-24] IPR002347 | 2.2e-39 | Glucose/ribitol dehydrogenase | ||
| [6-173] IPR002198 | 6.4e-23 | Short-chain dehydrogenase/reductase SDR | ||
| Orthology group | MCL23300 | Specific divergent | ||
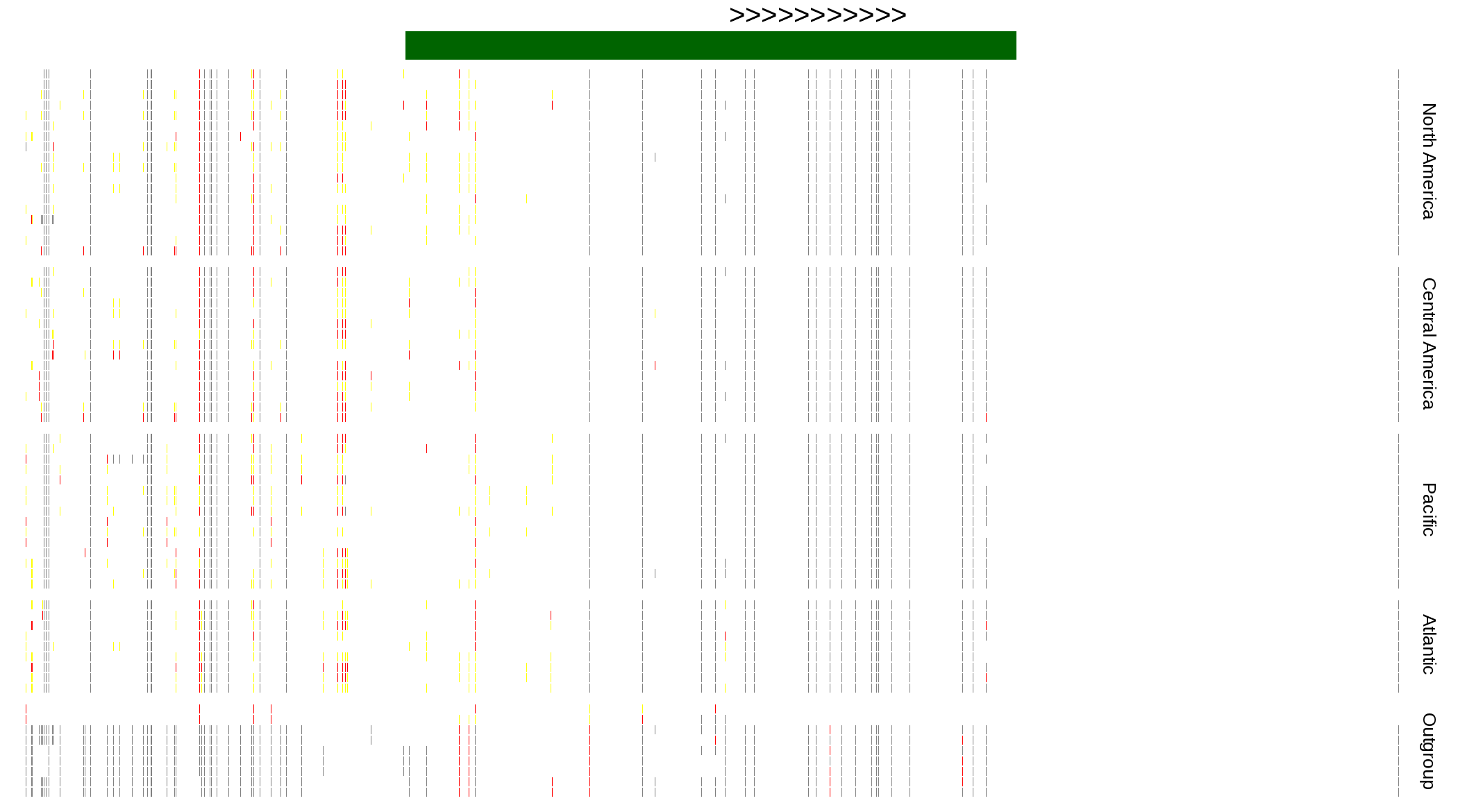
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS201627-TA
ATGAATTTTAAAAGTAAAGTCGTTGTCATAACGGGAGCTGCCTCTGGTATAGGAAAAGAAATTGCTATCAAGTTTGCTAAACTATCTGCTAATTTATCATTACTAGATATCGACTTAGAAAATTTGAAGTCCGTCGCAGAAGAGTGTCGTAATTTGAGTGAATCTAAAGTATTTCCATTGGTTGCAGATTTAGGAAAGAGTGATAATGTGAAAACTGTCGTTGATTTAATAAAACAGGAGTTCGGGAAGATTGATGTACTTGTGAATAGTGCTGGCATTGCTCGTATCGGTGATATAAAATGTGATGATTTATTGAAGGACGTTGACAATGTCCTTAATATTAATCTGATTTCTGGTATAACTCTTACACATTATGCTGCAGAGGCTTTAATAGAAAGTAAAGGATGTATTATTAATATAGCAAGTATAATGGCAGCGAAAGCAGGTATAGGTTCACTAGCGTACAATGTATCGAAGGCTGCAGTGTTACAATTTACAAAATGTGTGGCCTTGGAGCTGGCTGATAGCGGAGTTCGTGTGAATTGCATATCCCCAGGATCAGTGAAGACTAATGTTTTGAATAATGTGCTACCACCTTCAGAAGATTTCAATAAGTTTTGGGATGATTTAATTGAGGACCATCCAATGAGGCAGTTAACTACAACCGAAGATATCGCACACATGGCAGTGTTTTTGGCAACTGACAAAGCAAAAAGCATCACTGGCTGTAATTACATTGTGGACTCTGGAAAAACATTGATCAAATTTGCCTCTCACTTTCCATGA
>DPOGS201627-PA
MNFKSKVVVITGAASGIGKEIAIKFAKLSANLSLLDIDLENLKSVAEECRNLSESKVFPLVADLGKSDNVKTVVDLIKQEFGKIDVLVNSAGIARIGDIKCDDLLKDVDNVLNINLISGITLTHYAAEALIESKGCIINIASIMAAKAGIGSLAYNVSKAAVLQFTKCVALELADSGVRVNCISPGSVKTNVLNNVLPPSEDFNKFWDDLIEDHPMRQLTTTEDIAHMAVFLATDKAKSITGCNYIVDSGKTLIKFASHFP-