| DPOGS202043 | ||
|---|---|---|
| Transcript | DPOGS202043-TA | 306 bp |
| Protein | DPOGS202043-PA | 101 aa |
| Genomic position | DPSCF300053 + 97993-98666 | |
| RNAseq coverage | 2078x (Rank: top 6%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL009942 | 4e-51 | 89.90% | |
| Bombyx | BGIBMGA006752-TA | 1e-06 | 35.16% | |
| Drosophila | l(2)37Cg-PA | 5e-33 | 61.62% | |
| EBI UniRef50 | UniRef50_G6DPB7 | 2e-54 | 100.00% | Putative uncharacterized protein n=4 Tax=Pancrustacea RepID=G6DPB7_DANPL |
| NCBI RefSeq | XP_001604778.1 | 2e-32 | 60.61% | PREDICTED: similar to ENSANGP00000022241 [Nasonia vitripennis] |
| NCBI nr blastp | gi|345481326 | 2e-30 | 61.86% | PREDICTED: probable DNA-directed RNA polymerases I and III subunit RPAC2-like [Nasonia vitripennis] |
| NCBI nr blastx | gi|345481326 | 4e-29 | 61.86% | PREDICTED: probable DNA-directed RNA polymerases I and III subunit RPAC2-like [Nasonia vitripennis] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0003899 | 2.3e-24 | DNA-directed RNA polymerase activity | |
| GO:0003677 | 2.3e-24 | DNA binding | ||
| GO:0006351 | 2.3e-24 | transcription, DNA-dependent | ||
| KEGG pathway | nvi:100115834 | 6e-32 | ||
| K03020 (RPC19) | maps-> | Cytosolic DNA-sensing pathway | ||
| Purine metabolism | ||||
| Pyrimidine metabolism | ||||
| RNA polymerase | ||||
| InterPro domain | [16-100] IPR009025 | 2.3e-24 | DNA-directed RNA polymerase, RBP11-like | |
| Orthology group | MCL12650 | Single-copy universal gene | ||
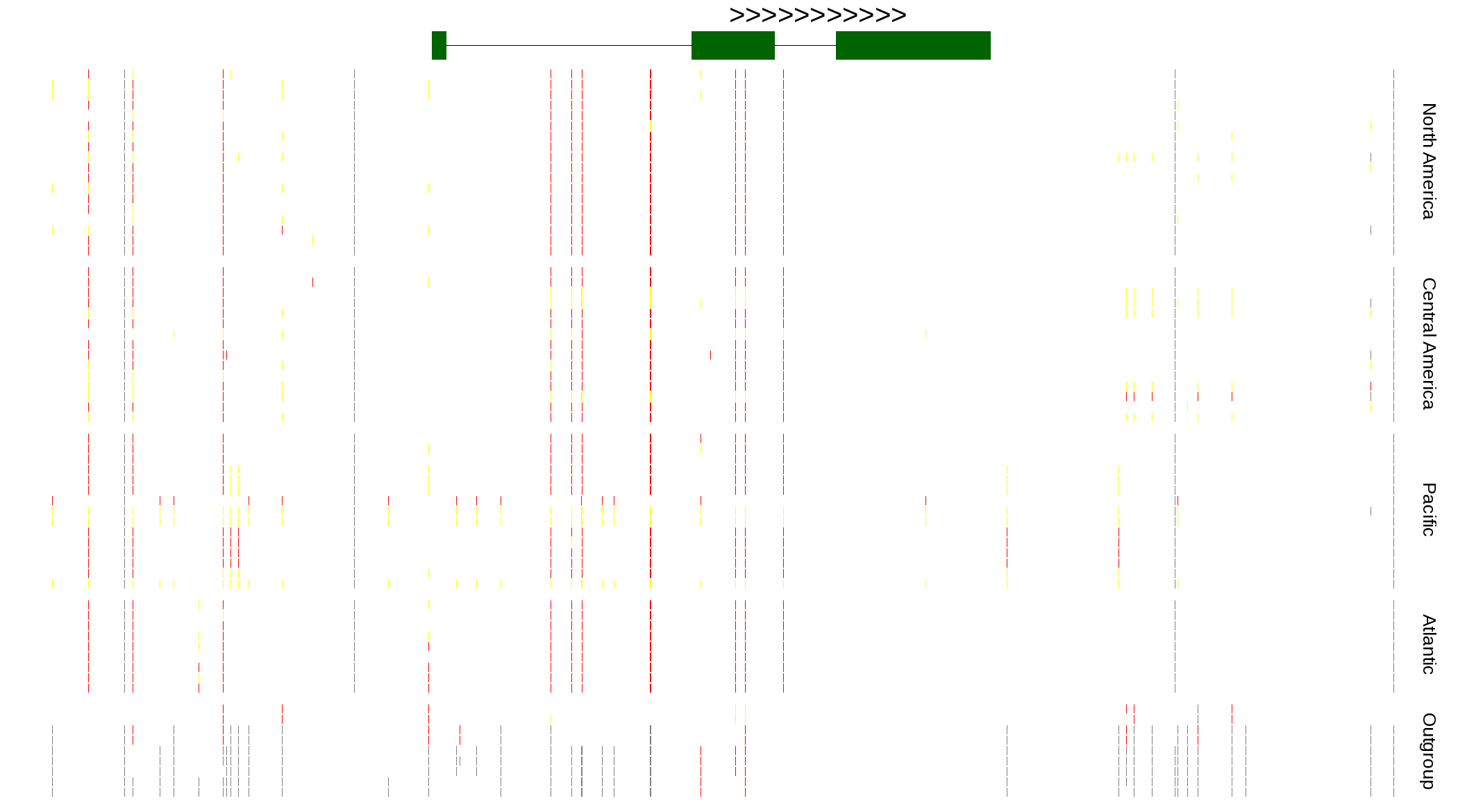
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS202043-TA
ATGATGAAAATATCTGAGTTGCCAGGTGACGAGGATAATGGATTGACCTGTAGAACTTACGTCTTTCAAAACGAAAGCCACACTTTAGGAAATGCTTTAAAATGCATCATTGACAGATATGAAGATGTCCAATTCTGTGGCTACACTGTCCCTCATCCAGCGGAAGCCAAGATGCACTTCAGGATCCAAACCCAAGAAACTCCCACATTAGATATACTGAAAAGAGGCCTAACAGACTTGCAAAAAGTCTGTGAACACACAATAGATTTATTTGAAAAGGAAGTAAAAGAATTTAGCAAACCTTAA
>DPOGS202043-PA
MMKISELPGDEDNGLTCRTYVFQNESHTLGNALKCIIDRYEDVQFCGYTVPHPAEAKMHFRIQTQETPTLDILKRGLTDLQKVCEHTIDLFEKEVKEFSKP-