| DPOGS202574 | ||
|---|---|---|
| Transcript | DPOGS202574-TA | 474 bp |
| Protein | DPOGS202574-PA | 157 aa |
| Genomic position | DPSCF300355 + 150187-151728 | |
| RNAseq coverage | 926x (Rank: top 14%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL013197 | 2e-79 | 89.81% | |
| Bombyx | BGIBMGA004278-TA | 7e-79 | 86.62% | |
| Drosophila | CG40042-PA | 3e-27 | 43.95% | |
| EBI UniRef50 | UniRef50_Q8MRW1 | 3e-25 | 43.95% | CG40042, isoform A n=20 Tax=Pancrustacea RepID=Q8MRW1_DROME |
| NCBI RefSeq | XP_001602964.1 | 3e-30 | 46.43% | PREDICTED: similar to ENSANGP00000012211 [Nasonia vitripennis] |
| NCBI nr blastp | gi|345480483 | 4e-29 | 46.43% | PREDICTED: mitochondrial import inner membrane translocase subunit Tim23-like [Nasonia vitripennis] |
| NCBI nr blastx | gi|321473076 | 6e-32 | 48.32% | hypothetical protein DAPPUDRAFT_194694 [Daphnia pulex] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0008565 | 1.9e-15 | protein transporter activity | |
| GO:0005743 | 1.9e-15 | mitochondrial inner membrane | ||
| GO:0015031 | 1.9e-15 | protein transport | ||
| KEGG pathway | ||||
| InterPro domain | [28-141] IPR003397 | 1.9e-15 | Mitochondrial inner membrane translocase complex, subunit Tim17/22 | |
| Orthology group | MCL12126 | Single-copy universal gene | ||
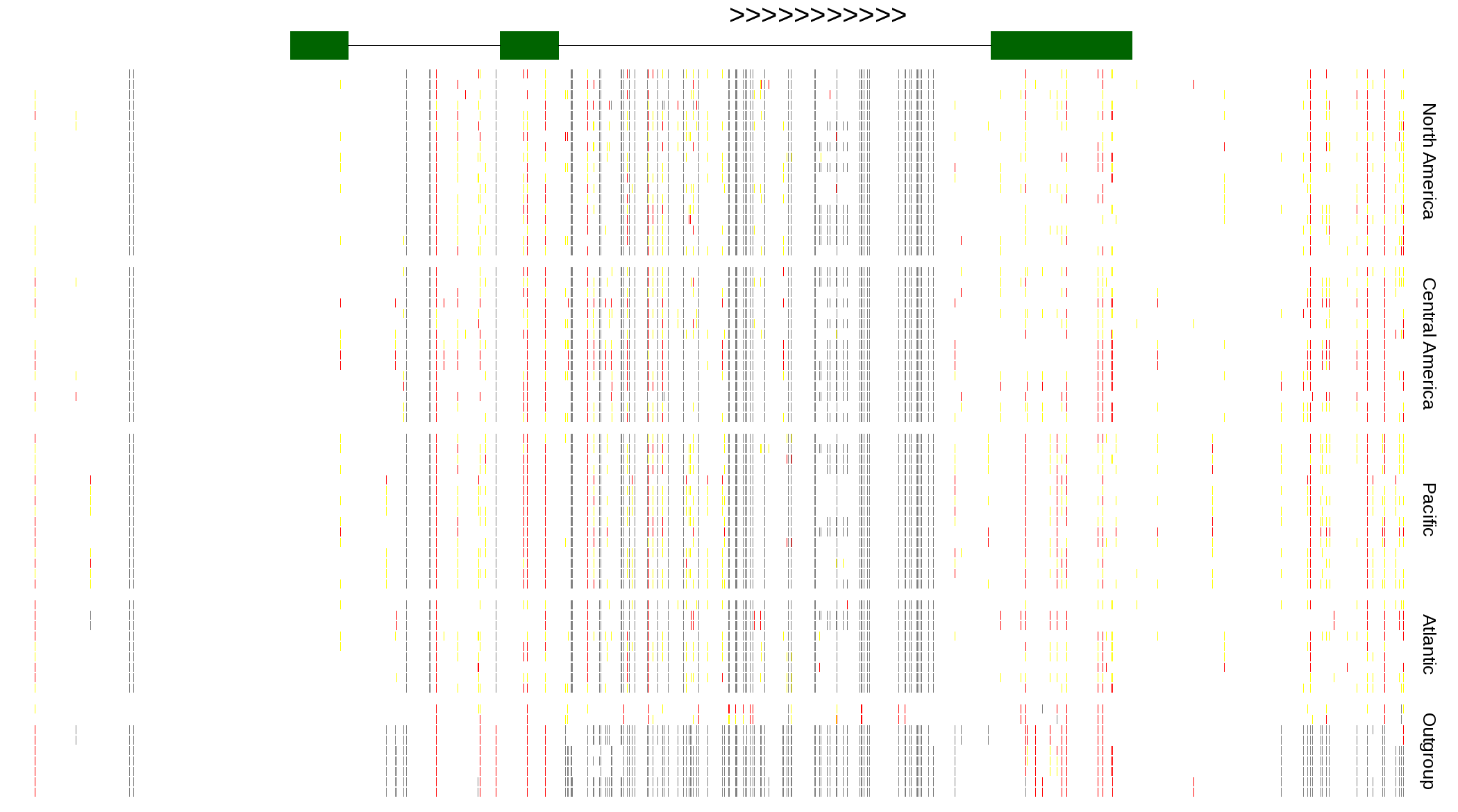
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS202574-TA
ATGCAACCAGAATTCCTTTATCCTGATGAGAGTCATATGGCTTCGACTGCGAGACGTTCAAATGTAGCTCTTCCTATTATCGGCATGTCCTTTATGACAGGATCAGGTATGGGAGGAATGGCAGGTCTCTACAAAGGATTACGAGCGACCACACTAGCTGGACAGACTGGCAAAGTCCGCCGAACACAACTAATTAATTATATAATGAAACAGGGTACTACGACTGGGTGTACCCTTGGTATTCTCGCGTCCTTCTACTCAACCCTGGCCCTAGGAGTCACCTGGTTACGAGACCAGGAGGACACAGCCAACACATTTATAGCGGCAACCGCTACCGGCATGTTGTACAAGAGTACCTCTGGACTCCGTTCTATGGGCCTGGGCGCTGTTGCTGGCCTCACCTTAGCAGGGATATACACATTGGTCACCGATAATGATAATATTTGGAGTAAGGCGAGATATATAAGAATGTGA
>DPOGS202574-PA
MQPEFLYPDESHMASTARRSNVALPIIGMSFMTGSGMGGMAGLYKGLRATTLAGQTGKVRRTQLINYIMKQGTTTGCTLGILASFYSTLALGVTWLRDQEDTANTFIAATATGMLYKSTSGLRSMGLGAVAGLTLAGIYTLVTDNDNIWSKARYIRM-