| DPOGS202653 | ||
|---|---|---|
| Transcript | DPOGS202653-TA | 213 bp |
| Protein | DPOGS202653-PA | 70 aa |
| Genomic position | DPSCF300039 - 169381-169593 | |
| RNAseq coverage | 0x (Rank: top 96%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL002223 | 5e-24 | 72.86% | |
| Bombyx | BGIBMGA000851-TA | 2e-20 | 58.57% | |
| Drosophila | CG10092-PA | 3e-08 | 37.68% | |
| EBI UniRef50 | UniRef50_E2ATT0 | 5e-13 | 50.00% | Probable arginyl-tRNA synthetase, mitochondrial n=6 Tax=Formicidae RepID=E2ATT0_CAMFO |
| NCBI RefSeq | XP_966449.1 | 2e-12 | 48.57% | PREDICTED: similar to arginyl-tRNA synthetase [Tribolium castaneum] |
| NCBI nr blastp | gi|307171176 | 2e-12 | 50.00% | Probable arginyl-tRNA synthetase, mitochondrial [Camponotus floridanus] |
| NCBI nr blastx | gi|307171176 | 9e-12 | 50.00% | Probable arginyl-tRNA synthetase, mitochondrial [Camponotus floridanus] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0004814 | 1.6e-12 | arginine-tRNA ligase activity | |
| GO:0005524 | 1.6e-12 | ATP binding | ||
| GO:0006420 | 1.6e-12 | arginyl-tRNA aminoacylation | ||
| GO:0005737 | 4.1e-12 | cytoplasm | ||
| GO:0006418 | 7.2e-12 | tRNA aminoacylation for protein translation | ||
| GO:0000166 | 7.2e-12 | nucleotide binding | ||
| GO:0004812 | 7.2e-12 | aminoacyl-tRNA ligase activity | ||
| KEGG pathway | tca:654913 | 5e-12 | ||
| K01887 (RARS, argS) | maps-> | Aminoacyl-tRNA biosynthesis | ||
| InterPro domain | [2-70] IPR008909 | 1.6e-12 | DALR anticodon binding | |
| [9-70] IPR001278 | 4.1e-12 | Arginyl-tRNA synthetase, class Ia | ||
| [1-70] IPR009080 | 7.2e-12 | Aminoacyl-tRNA synthetase, class 1a, anticodon-binding | ||
| Orthology group | ||||
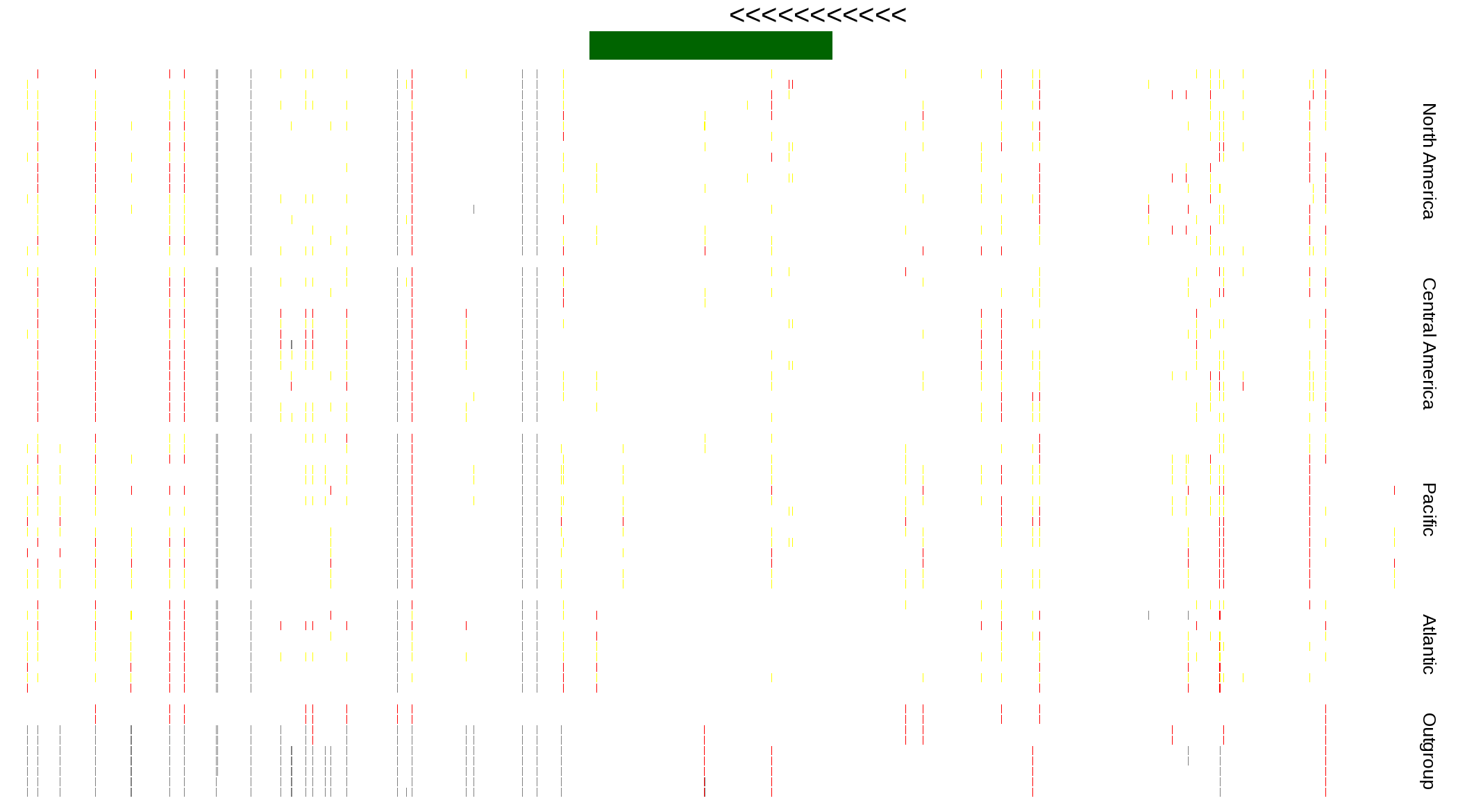
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS202653-TA
ATGAACCAGGCATATAATGAAAGTGAAGCTTGTATAATAGTAAATTATTTATTTCGACTATCAAATATGGTCAATCGTATGTTCAATGAGCTGAAAGTTAAAAATGTTAATCGTGATGTCGCTTCCCAAAGGCTCCTCGTTTTTAATTCTGCAAGATTTGTAATTAAAACAGCTTTGGAAATTCTAGGTGTAAAACCTCTACTTGAAATGTAA
>DPOGS202653-PA
MNQAYNESEACIIVNYLFRLSNMVNRMFNELKVKNVNRDVASQRLLVFNSARFVIKTALEILGVKPLLEM-