| DPOGS202851 | ||
|---|---|---|
| Transcript | DPOGS202851-TA | 729 bp |
| Protein | DPOGS202851-PA | 242 aa |
| Genomic position | DPSCF300018 + 1098424-1099503 | |
| RNAseq coverage | 45x (Rank: top 71%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL002691 | 1e-110 | 93.13% | |
| Bombyx | BGIBMGA010490-TA | 4e-110 | 91.45% | |
| Drosophila | slp2-PA | 6e-61 | 67.05% | |
| EBI UniRef50 | UniRef50_F4X2Y0 | 1e-66 | 63.37% | Fork head domain transcription factor slp2 n=8 Tax=Formicidae RepID=F4X2Y0_ACREC |
| NCBI RefSeq | XP_001120632.1 | 2e-65 | 62.98% | PREDICTED: similar to sloppy paired 2 CG2939-PA [Apis mellifera] |
| NCBI nr blastp | gi|332018620 | 5e-66 | 63.37% | Fork head domain transcription factor slp2 [Acromyrmex echinatior] |
| NCBI nr blastx | gi|350399880 | 9e-67 | 54.81% | PREDICTED: fork head domain transcription factor slp2-like [Bombus impatiens] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0006355 | 5.2e-61 | regulation of transcription, DNA-dependent | |
| GO:0043565 | 5.2e-61 | sequence-specific DNA binding | ||
| GO:0003700 | 5.2e-61 | sequence-specific DNA binding transcription factor activity | ||
| KEGG pathway | ||||
| InterPro domain | [47-137] IPR001766 | 5.2e-61 | Transcription factor, fork head | |
| [42-142] IPR011991 | 3.1e-45 | Winged helix-turn-helix transcription repressor DNA-binding | ||
| Orthology group | MCL15946 | Insect specific | ||
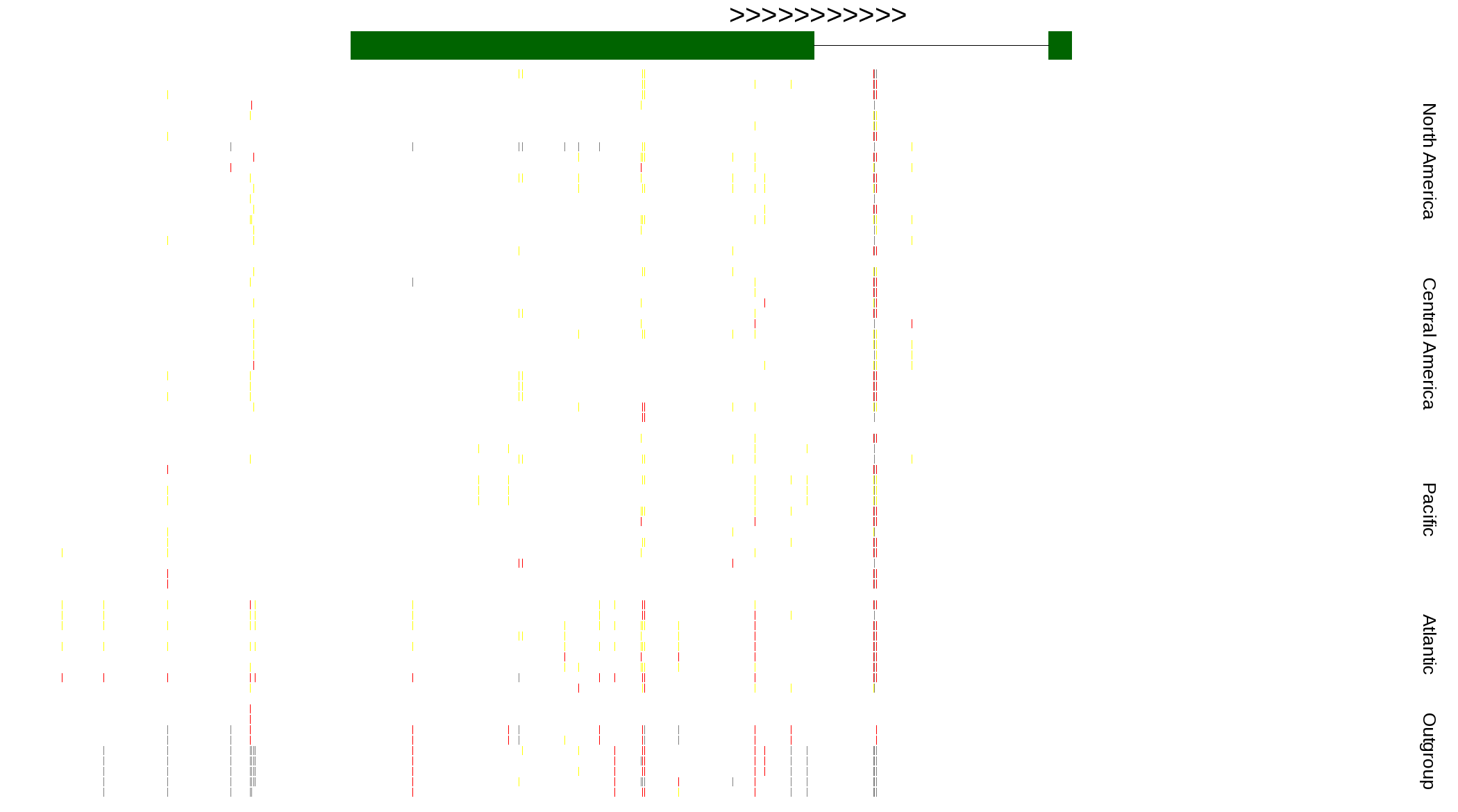
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS202851-TA
ATGAGTCCTCCGAGCTTCGCCATCCGAGACCTTCTGCCGGAGGCGTCGTCGCGGTCCCCCTCGCCCTCCGACACGGAGCTGGATGTCACCGGTACGGAGACGCCGCCGCCAGGCTGCTCCACCACCGATGAAAAGAAAAATGAAAAACCGGCCTACAGCTACAATGCGCTCATCATGATGGCCATACGCAGCAGTCCCGAGAGACGGCTCACGCTCAACGGCATCTATGAGTACATCATGAAGAACTTCCCCTACTACAAGGAGAACAAGCAGGGCTGGCAGAACTCTATTCGACACAATCTGAGTTTGAATAAGTGTTTCGTGAAAGTCCCGCGTCACTACGACGATCCCGGGAAAGGGAACTACTGGATGCTGGACCCTTCGTCTGACGACGTCTTCATCGGTGGCACCACTGGCAAGCTGAGGAGGAGATCCACCGCCGCATCTAGGTCTCGGCTCGCAGCCTTCAAGAGAGGTGCCGTCCTCCACCCCATGTTCGGGAATCCGTATGCGTCTCTGGTGGGTCTGTACCCACCCTTACTATCGGTGTACGGGAGGTACGCCCTGCCGAGCCCGCTGCTGAGGTTGCCGCCGCCTCCTCCCAGTCCCTATCACCAGCTGTATCGCAGGATCCACGGCCTGTCACCTCCCGGCAGCCCACCAGCGGAGCCGGAGTCACATCTGATGAAGCACACTTGTCTGAATTATGACAAATTAGGGTTGCAGTAA
>DPOGS202851-PA
MSPPSFAIRDLLPEASSRSPSPSDTELDVTGTETPPPGCSTTDEKKNEKPAYSYNALIMMAIRSSPERRLTLNGIYEYIMKNFPYYKENKQGWQNSIRHNLSLNKCFVKVPRHYDDPGKGNYWMLDPSSDDVFIGGTTGKLRRRSTAASRSRLAAFKRGAVLHPMFGNPYASLVGLYPPLLSVYGRYALPSPLLRLPPPPPSPYHQLYRRIHGLSPPGSPPAEPESHLMKHTCLNYDKLGLQ-