| DPOGS203462 | ||
|---|---|---|
| Transcript | DPOGS203462-TA | 297 bp |
| Protein | DPOGS203462-PA | 98 aa |
| Genomic position | DPSCF301835 - 93-714 | |
| RNAseq coverage | 161x (Rank: top 52%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL014138 | 1e-45 | 87.10% | |
| Bombyx | BGIBMGA003564-TA | 1e-45 | 88.17% | |
| Drosophila | Oat-PA | 8e-38 | 70.97% | |
| EBI UniRef50 | UniRef50_F8W5L3 | 1e-36 | 67.74% | Uncharacterized protein n=1 Tax=Danio rerio RepID=F8W5L3_DANRE |
| NCBI RefSeq | XP_001650687.1 | 2e-38 | 69.89% | ornithine aminotransferase [Aedes aegypti] |
| NCBI nr blastp | gi|317419517 | 3e-37 | 67.74% | Ornithine aminotransferase, mitochondrial [Dicentrarchus labrax] |
| NCBI nr blastx | gi|348508574 | 2e-35 | 69.89% | PREDICTED: ornithine aminotransferase, mitochondrial-like [Oreochromis niloticus] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0008483 | 6e-40 | transaminase activity | |
| GO:0030170 | 6e-40 | pyridoxal phosphate binding | ||
| GO:0003824 | 3.3e-17 | catalytic activity | ||
| KEGG pathway | aag:AaeL_AAEL005289 | 8e-38 | ||
| K00819 (E2.6.1.13, rocD) | maps-> | Arginine and proline metabolism | ||
| InterPro domain | [1-90] IPR010164 | 6e-40 | Ornithine aminotransferase | |
| [1-90] IPR005814 | 6e-40 | Aminotransferase class-III | ||
| [2-90] IPR015421 | 3.3e-17 | Pyridoxal phosphate-dependent transferase, major region, subdomain 1 | ||
| [1-90] IPR015424 | 1.8e-14 | Pyridoxal phosphate-dependent transferase, major domain | ||
| Orthology group | ||||
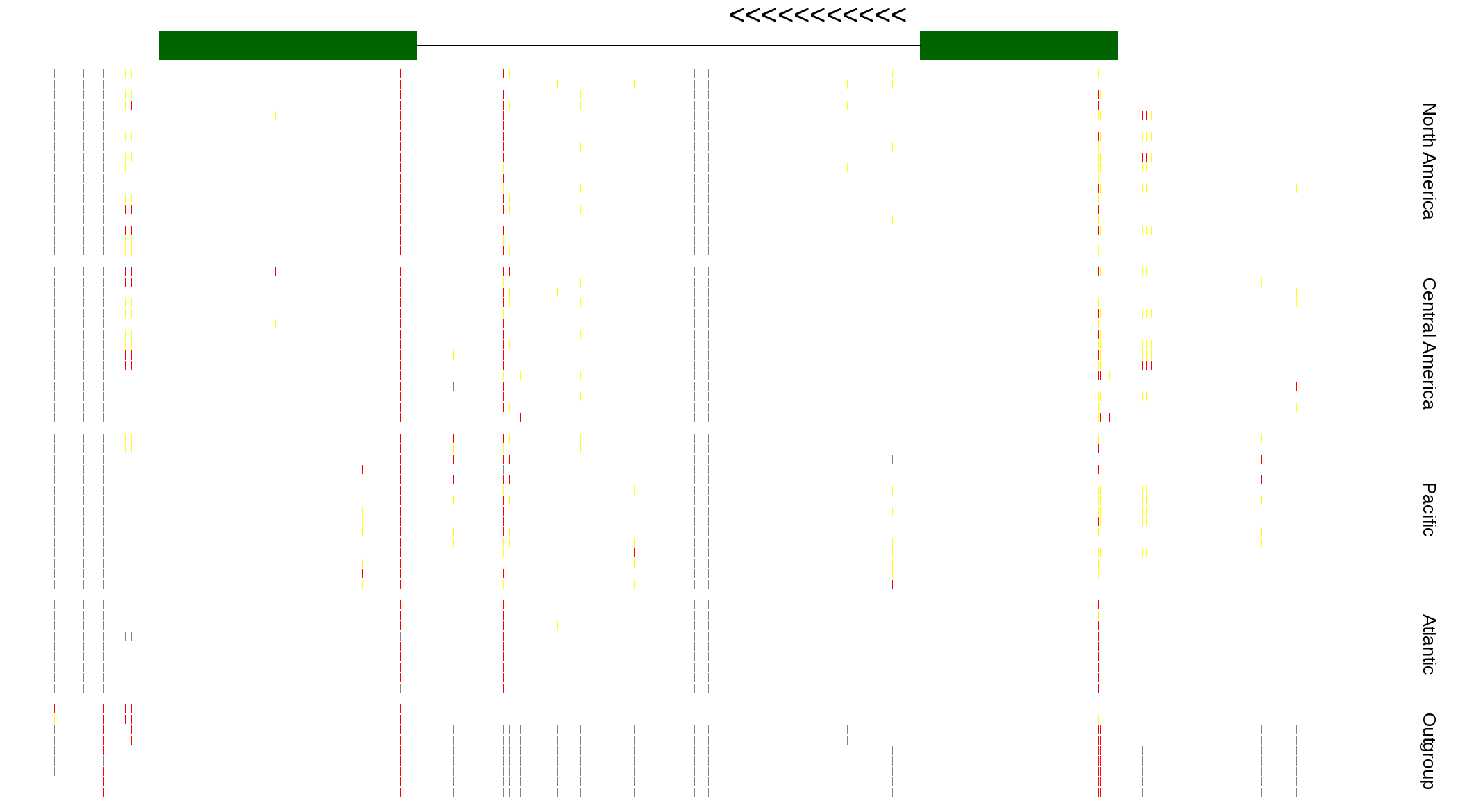
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS203462-TA
ATGACAGAGCTACTAGGCTATGATCGTCTGCTGCCAATGAACACAGGAGTTGAAGGTGGGGAGAGCGCTTGCAAGATAGCCAGGAAATGGGGATATGACGTCAAGAAAATACCAAAAGATCAGGCAAAGATAATTTTCGCTGAGAACAACTTCTGGGGTCGCACGTTGTCCGCGGTGTCATCTTCGTCGGACCCCACTTGCTACGAGGGTTTCGGTCCTTACATGCCGTGTTTCCAGATGATACCCTACAATAATATACCTGCTCTAGAGGTCGGATTGTGTTACACATATATTTAA
>DPOGS203462-PA
MTELLGYDRLLPMNTGVEGGESACKIARKWGYDVKKIPKDQAKIIFAENNFWGRTLSAVSSSSDPTCYEGFGPYMPCFQMIPYNNIPALEVGLCYTYI-