| DPOGS203863 | ||
|---|---|---|
| Transcript | DPOGS203863-TA | 303 bp |
| Protein | DPOGS203863-PA | 100 aa |
| Genomic position | DPSCF300010 + 3598380-3598876 | |
| RNAseq coverage | 106x (Rank: top 60%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL008486 | 5e-10 | 65.91% | |
| Bombyx | % | |||
| Drosophila | Vha36-3-PA | 2e-07 | 51.16% | |
| EBI UniRef50 | UniRef50_D0A289 | 5e-07 | 53.57% | Vacuolar ATP synthase subunit d, putative n=6 Tax=Trypanosomatidae RepID=D0A289_TRYB9 |
| NCBI RefSeq | XP_001600508.1 | 2e-07 | 40.85% | PREDICTED: similar to vacuolar ATP synthase subunit D [Nasonia vitripennis] |
| NCBI nr blastp | gi|322495317 | 7e-07 | 50.79% | putative vacuolar ATP synthase subunit d [Leishmania mexicana MHOM/GT/2001/U1103] |
| NCBI nr blastx | gi|358059294 | 2e-10 | 43.42% | hypothetical protein E5Q_01637 [Mixia osmundae IAM 14324] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0033178 | 9.9e-15 | proton-transporting two-sector ATPase complex, catalytic domain | |
| GO:0015991 | 9.9e-15 | ATP hydrolysis coupled proton transport | ||
| GO:0042626 | 9.9e-15 | ATPase activity, coupled to transmembrane movement of substances | ||
| GO:0046961 | 9.9e-15 | proton-transporting ATPase activity, rotational mechanism | ||
| KEGG pathway | lma:LmjF35.0700 | 1e-07 | ||
| K02149 (ATPeVD, ATP6M) | maps-> | Collecting duct acid secretion | ||
| Oxidative phosphorylation | ||||
| Phagosome | ||||
| Vibrio cholerae infection | ||||
| Epithelial cell signaling in Helicobacter pylori infection | ||||
| InterPro domain | [6-51] IPR002699 | 9.9e-15 | ATPase, V1/A1 complex, subunit D | |
| Orthology group | ||||
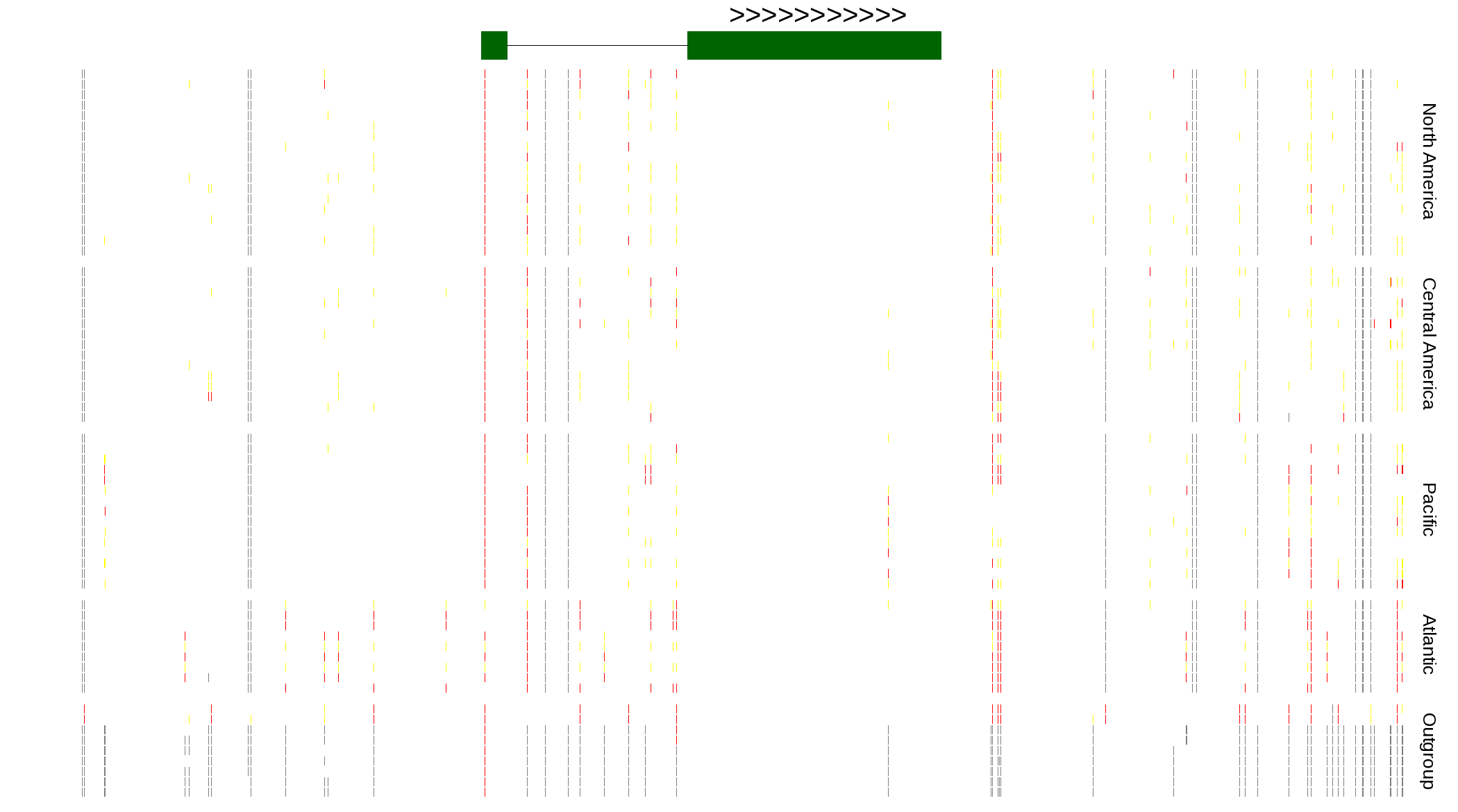
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS203863-TA
ATGGGTTGTAATAAAAGGAGATATTCTAAGAAAGCGAACGTCCTGGGTAAAGTCGTGATACCGAGACTGAATGTAACTACGGATTATATAAACTCTGAACTAGAAGAGAAGGAGAGAGAGGAAGTTTTCCGTCTAAAGAGAGTTAAACAGAAGAAAGACTTTGATAAGAAGCAGGAAGAGAATAAGAGGAGGCAGAACGACGCCGATAAGAAGGCTCGCGAAAAAGGAGGCCTCCCTGATGACGACGAAGACCCGAAACAAGCTTCGCCAACCCCAGCGCCACCAGTCCCTCAAGATTCTTGA
>DPOGS203863-PA
MGCNKRRYSKKANVLGKVVIPRLNVTTDYINSELEEKEREEVFRLKRVKQKKDFDKKQEENKRRQNDADKKAREKGGLPDDDEDPKQASPTPAPPVPQDS-