| DPOGS204032 | ||
|---|---|---|
| Transcript | DPOGS204032-TA | 570 bp |
| Protein | DPOGS204032-PA | 189 aa |
| Genomic position | DPSCF300138 + 18951-21904 | |
| RNAseq coverage | 611x (Rank: top 21%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL008165 | 1e-35 | 91.78% | |
| Bombyx | BGIBMGA004781-TA | 7e-69 | 80.92% | |
| Drosophila | Plip-PB | 8e-76 | 67.71% | |
| EBI UniRef50 | UniRef50_Q86BN8 | 9e-74 | 67.71% | Protein-tyrosine phosphatase mitochondrial 1-like protein n=35 Tax=Coelomata RepID=PTPM1_DROME |
| NCBI RefSeq | XP_001358634.2 | 1e-76 | 68.75% | GA10281 [Drosophila pseudoobscura pseudoobscura] |
| NCBI nr blastp | gi|198452108 | 2e-75 | 68.75% | GA10281 [Drosophila pseudoobscura pseudoobscura] |
| NCBI nr blastx | gi|307212581 | 3e-72 | 67.19% | Protein-tyrosine phosphatase mitochondrial 1-like protein [Harpegnathos saltator] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0008138 | 2.3e-19 | protein tyrosine/serine/threonine phosphatase activity | |
| GO:0006470 | 2.3e-19 | protein dephosphorylation | ||
| KEGG pathway | ||||
| InterPro domain | [48-179] IPR000340 | 2.3e-19 | Dual specificity phosphatase, catalytic domain | |
| [28-185] IPR020422 | 2.6e-11 | Dual specificity phosphatase, subgroup, catalytic domain | ||
| Orthology group | MCL11892 | Single-copy universal gene | ||
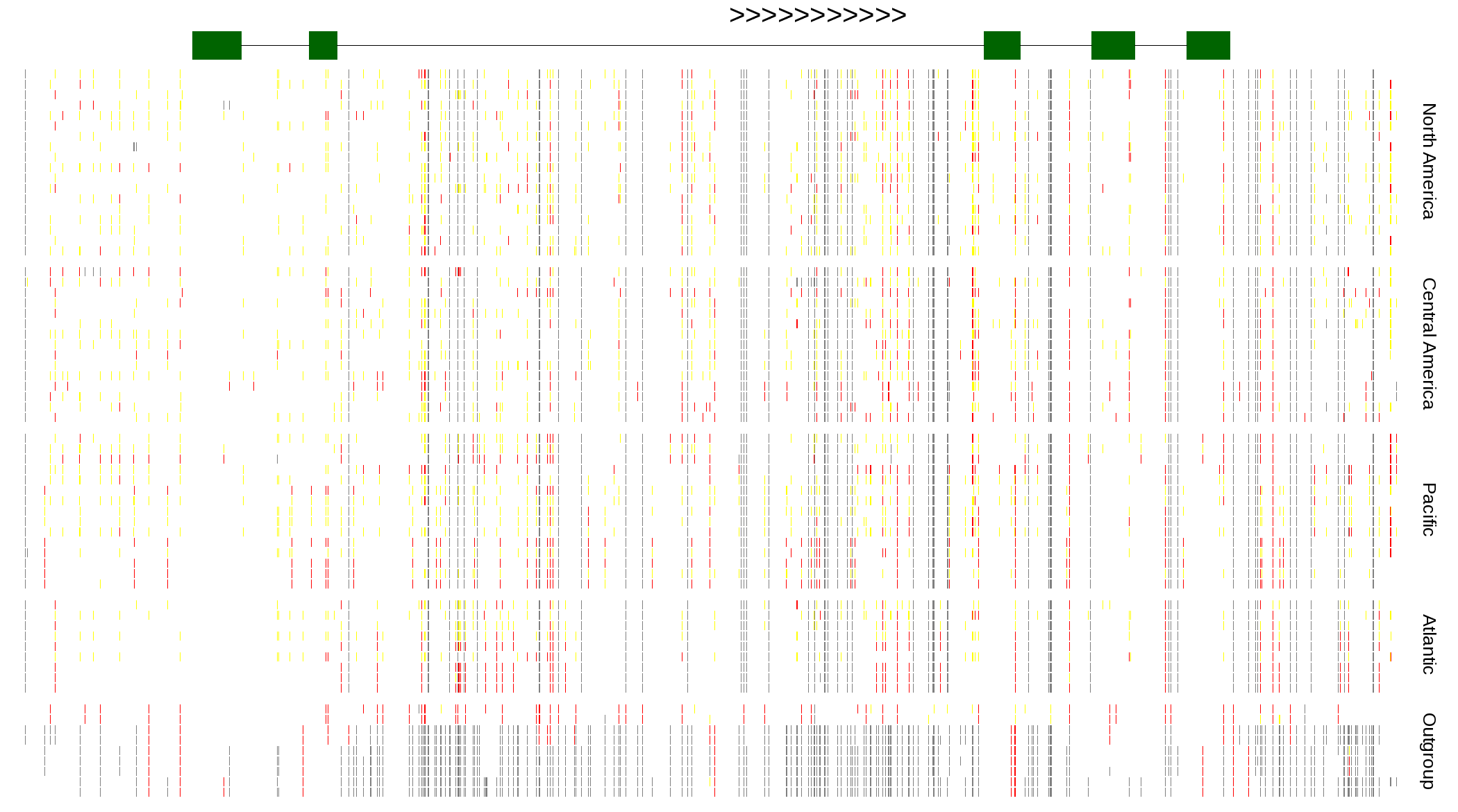
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS204032-TA
ATGTTTGCGAGAGTGACCTTCTATCCGACTCTGTTGTATAATGTGCTGATGGAAAAGGTGACCAGCAGACGATGGTACGACAGGATGGACGATACAGTGATCCTAGGAGCTCTGCCCTTTCAAGGAATGACCAAACAATTAATAGAAGATGAAAACATTAAAGGAGTTGTTTCAATGAATGAGACATACGAATTAAAAATATTTTCCAATGATGGCGAGAAATGGCGGGAACACGGTGTAGAATTTCTTCAGCTGGCAACAACAGATATCTTCGAAGCACCTGACCAAGACAAACTCATTGAAGGCGTGCGGTTTATTAACAGATTTCTCCCTCGATCATCACAAAGCCTCTCAACCAGTGATGAACGGACTCGCGGAACCGTGTACGTGCACTGCAAGGCTGGTCGCACGAGGAGTGCCACTCTGGTCGGGTGCTACCTCATGATGAGAAACGGCTGGTCTCCAAACGAGGCTGTAGACTACATGAAGTCGCGCCGGCCACACATACTCCTACACACGAAGCAATGGCAGGCGCTAGATATATTTTACAAGAGACATGTCAAGACATAG
>DPOGS204032-PA
MFARVTFYPTLLYNVLMEKVTSRRWYDRMDDTVILGALPFQGMTKQLIEDENIKGVVSMNETYELKIFSNDGEKWREHGVEFLQLATTDIFEAPDQDKLIEGVRFINRFLPRSSQSLSTSDERTRGTVYVHCKAGRTRSATLVGCYLMMRNGWSPNEAVDYMKSRRPHILLHTKQWQALDIFYKRHVKT-