| DPOGS204176 | ||
|---|---|---|
| Transcript | DPOGS204176-TA | 690 bp |
| Protein | DPOGS204176-PA | 229 aa |
| Genomic position | DPSCF300034 - 25399-28553 | |
| RNAseq coverage | 4x (Rank: top 89%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL007771 | 1e-111 | 84.96% | |
| Bombyx | BGIBMGA005091-TA | 2e-116 | 88.00% | |
| Drosophila | amon-PA | 4e-96 | 74.55% | |
| EBI UniRef50 | UniRef50_G5ECN9 | 1e-75 | 57.85% | Prohormone convertase 2 n=17 Tax=Bilateria RepID=G5ECN9_CAEEL |
| NCBI RefSeq | XP_308012.4 | 1e-99 | 75.56% | AGAP002176-PA [Anopheles gambiae str. PEST] |
| NCBI nr blastp | gi|347967315 | 1e-98 | 75.56% | AGAP002176-PA [Anopheles gambiae str. PEST] |
| NCBI nr blastx | gi|170053889 | 2e-98 | 76.47% | neuroendocrine convertase [Culex quinquefasciatus] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0004252 | 7.2e-30 | serine-type endopeptidase activity | |
| GO:0006508 | 7.2e-30 | proteolysis | ||
| KEGG pathway | ||||
| InterPro domain | [5-228] IPR015500 | 3.9e-96 | Peptidase S8, subtilisin-related | |
| [58-204] IPR008979 | 3.4e-46 | Galactose-binding domain-like | ||
| [109-195] IPR002884 | 7.2e-30 | Proprotein convertase, P | ||
| [5-65] IPR000209 | 2.5e-08 | Peptidase S8/S53, subtilisin/kexin/sedolisin | ||
| Orthology group | MCL30299 | Lepidoptera specific | ||
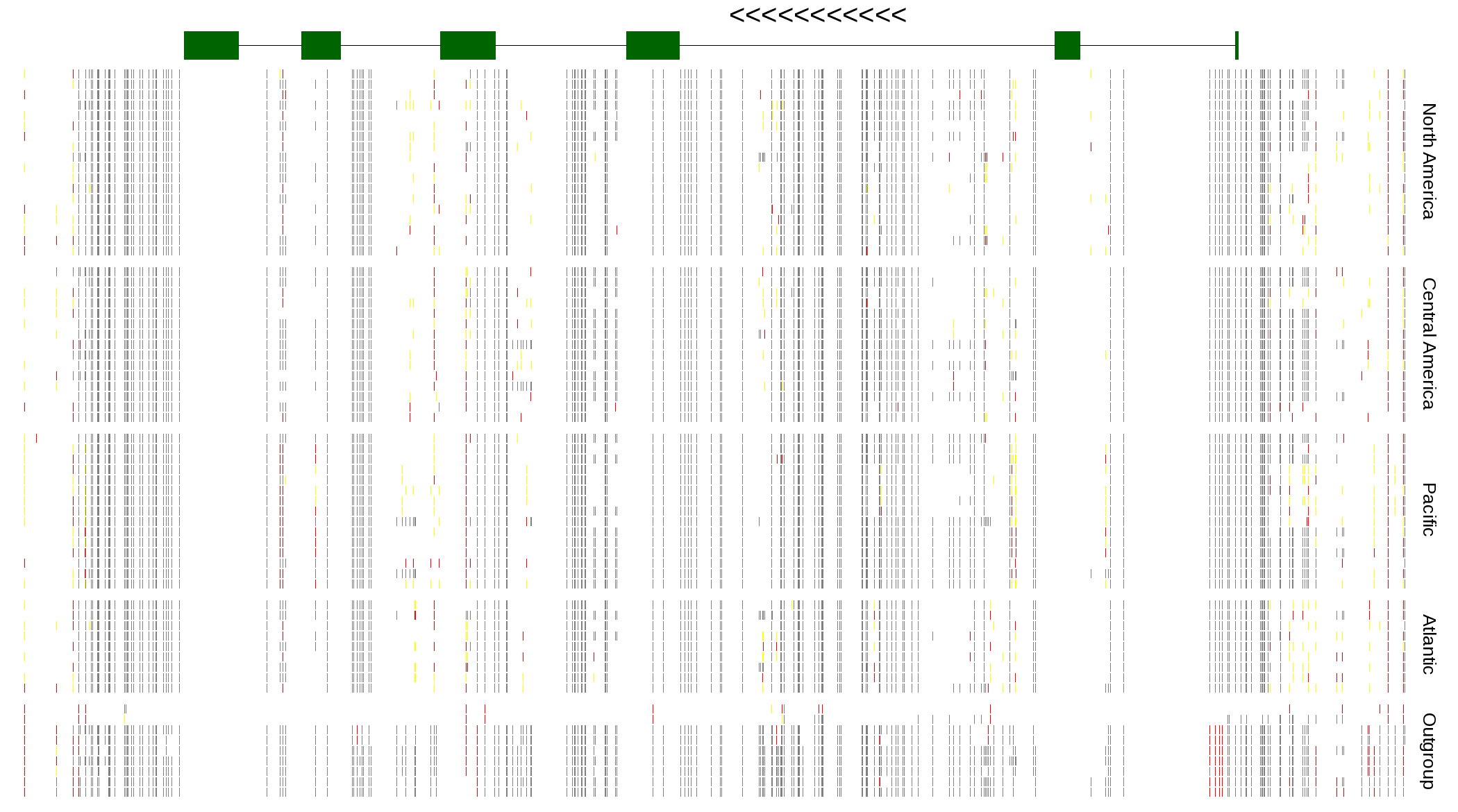
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS204176-TA
ATGTTGAAAGTTCCTAACCTGACATGGCGTGACATTCAACATCTCACTGTTTTGACATCAAAGAGGAACTCATTGTATGACGCGAAGGGCCGTTTCCATTGGACTATGAACGGAGTGGGTCTGGAATTCAATCATCTGTTTGGTTTCGGTGTCCTTGACGCCGGAGCTATGACAGCACTCGCCGCAAATTGGCGATCTGTGCCGCCGAGATACCATTGCGAAGCCGGCTCAGTCGACACTCACACCGAACTCCCATCGGAAGGCAGTATCACACTCCGGATAGACACGTCAGCATGCGCGGGCACCCCTAGCGAGGTTCGGTATTTGGAGCACGTGCAAGCTGTTGTCAGTGCCAACGCTACCAGACGAGGGGACTTGGAACTCTTCCTCACCAGCCCTATGGGAACCAAATCGATGATCCTGAGCAGGCGTGCAAATGATGACGACAGCCGTGACGGTTTTACAAAGTGGCCGTTCATGACTACGCACACTTGGGGAGAGTATCCGCAAGGAGTTTGGTCTTTGGAGGCTAGATTCAGCAGCCCAGGTCGATCTGGTTGGTTGCGAGGCTGGTCACTGGTGCTCCATGGCACACGAGCGCCGCCATACGCTCAGCTCCAGCCGCAAGACCCTCGCTCCAAGCTAGCAGTCGTCAAGAAGGCACACGAAGACAACGCTATCAACGACTAG
>DPOGS204176-PA
MLKVPNLTWRDIQHLTVLTSKRNSLYDAKGRFHWTMNGVGLEFNHLFGFGVLDAGAMTALAANWRSVPPRYHCEAGSVDTHTELPSEGSITLRIDTSACAGTPSEVRYLEHVQAVVSANATRRGDLELFLTSPMGTKSMILSRRANDDDSRDGFTKWPFMTTHTWGEYPQGVWSLEARFSSPGRSGWLRGWSLVLHGTRAPPYAQLQPQDPRSKLAVVKKAHEDNAIND-