| DPOGS204203 | ||
|---|---|---|
| Transcript | DPOGS204203-TA | 582 bp |
| Protein | DPOGS204203-PA | 193 aa |
| Genomic position | DPSCF300034 + 596718-598512 | |
| RNAseq coverage | 971x (Rank: top 13%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL004123 | 8e-93 | 87.05% | |
| Bombyx | BGIBMGA005161-TA | 1e-95 | 83.94% | |
| Drosophila | CG1969-PD | 7e-78 | 75.28% | |
| EBI UniRef50 | UniRef50_Q9VAI0 | 4e-76 | 75.28% | Probable glucosamine 6-phosphate N-acetyltransferase n=12 Tax=Sophophora RepID=GNA1_DROME |
| NCBI RefSeq | NP_001040128.1 | 4e-94 | 83.94% | glucosamine-6-phosphate N-acetyltransferase [Bombyx mori] |
| NCBI nr blastp | gi|114052422 | 6e-93 | 83.94% | glucosamine-6-phosphate N-acetyltransferase [Bombyx mori] |
| NCBI nr blastx | gi|114052422 | 8e-90 | 83.94% | glucosamine-6-phosphate N-acetyltransferase [Bombyx mori] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0008152 | 1.6e-08 | metabolic process | |
| GO:0008080 | 1.6e-08 | N-acetyltransferase activity | ||
| KEGG pathway | dan:Dana_GF23369 | 1e-77 | ||
| K00621 (E2.3.1.4, GNA1) | maps-> | Amino sugar and nucleotide sugar metabolism | ||
| InterPro domain | [49-181] IPR016181 | 1.1e-29 | Acyl-CoA N-acyltransferase | |
| [106-181] IPR000182 | 1.6e-08 | GCN5-related N-acetyltransferase (GNAT) domain | ||
| Orthology group | MCL13006 | Single-copy universal gene | ||
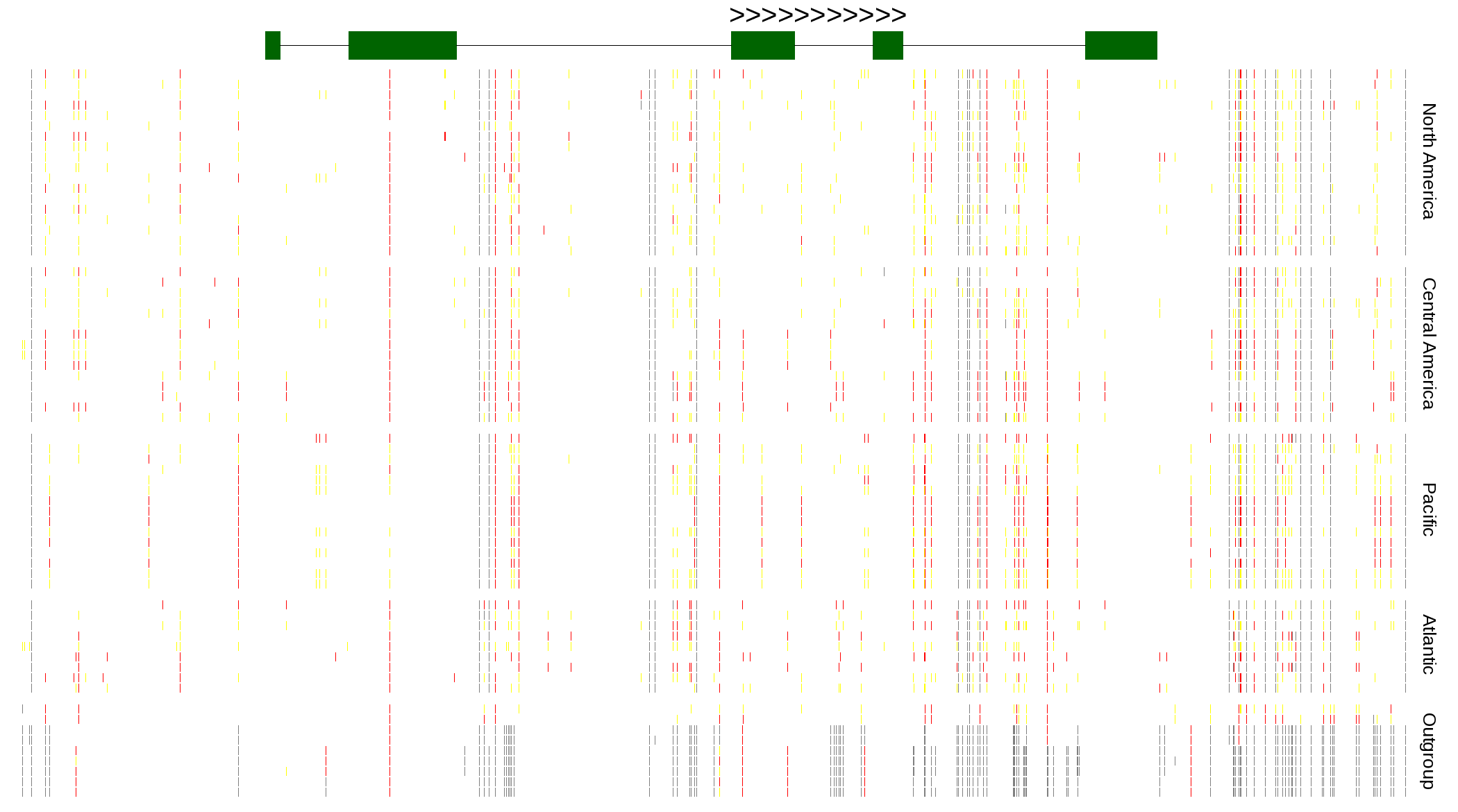
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS204203-TA
ATGGGCATGGAAAAGCAACATAGTATGGAGGACGAAAATTCGGAATATCTTTATCATCCAGATATACTTAAAAAATTGGATTTTGGCAAGAGCCCAGCCAAATTTAATCCCTGCATCAGTGCAGCTCATCCAGGTGAAGATTGGATGGTTGTGAGACCGCTTCAAAGAGCTGACTATGATAAGGGATTTCTCCAGCTTCTCAGTCAGTTAACCAGTACTGGTAGCATATCACGGAAGCAATTTGATGAACGTTTTACACAGATGAAAAGCGCTGGCGGATATTACGTGACAGTTATAGAGGACAAGCGTATTTCAAAAATAATTGGAGCTGCAACACTAACAATAGAACAAAAATTTATTCATAATTGCTCTGTGAGAGGCCGCTTAGAGGATGTAGTTGTGAATGACACTTATCGGGGAAAACAATTAGGAAAACTAATTGTAGTGACTGTATCATTACTGGCACAAGAACTGGGTTGTTATAAGATGTCATTAGACTGTAAAGACAAACTGATCAAGTTCTATGAAAGCCTCGGTTATAAACTTGAGCCCGGTAACTCTAACGCTATGAATATGAGGTAA
>DPOGS204203-PA
MGMEKQHSMEDENSEYLYHPDILKKLDFGKSPAKFNPCISAAHPGEDWMVVRPLQRADYDKGFLQLLSQLTSTGSISRKQFDERFTQMKSAGGYYVTVIEDKRISKIIGAATLTIEQKFIHNCSVRGRLEDVVVNDTYRGKQLGKLIVVTVSLLAQELGCYKMSLDCKDKLIKFYESLGYKLEPGNSNAMNMR-