| DPOGS204688 | ||
|---|---|---|
| Transcript | DPOGS204688-TA | 654 bp |
| Protein | DPOGS204688-PA | 217 aa |
| Genomic position | DPSCF300170 + 86647-88124 | |
| RNAseq coverage | 170x (Rank: top 51%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL009459 | 2e-81 | 63.59% | |
| Bombyx | BGIBMGA010237-TA | 9e-90 | 71.89% | |
| Drosophila | Gs1l-PB | 4e-54 | 50.23% | |
| EBI UniRef50 | UniRef50_F4X6L0 | 5e-62 | 55.81% | GS1-like protein n=29 Tax=cellular organisms RepID=F4X6L0_ACREC |
| NCBI RefSeq | XP_314275.4 | 7e-65 | 56.74% | AGAP003372-PA [Anopheles gambiae str. PEST] |
| NCBI nr blastp | gi|332030576 | 4e-66 | 58.33% | Haloacid dehalogenase-like hydrolase domain-containing protein 1A [Acromyrmex echinatior] |
| NCBI nr blastx | gi|332030576 | 1e-65 | 58.33% | Haloacid dehalogenase-like hydrolase domain-containing protein 1A [Acromyrmex echinatior] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0008152 | 1.1e-13 | metabolic process | |
| GO:0003824 | 1.1e-13 | catalytic activity | ||
| GO:0016787 | 1.1e-12 | hydrolase activity | ||
| KEGG pathway | ath:AT4G21470 | 4e-31 | ||
| K00861 (RFK, FMN1) | maps-> | Riboflavin metabolism | ||
| InterPro domain | [68-216] IPR023214 | 1.6e-36 | HAD-like domain | |
| [1-176] IPR005834 | 1.1e-13 | Haloacid dehalogenase-like hydrolase | ||
| [130-182] IPR006402 | 1.1e-12 | HAD-superfamily hydrolase, subfamily IA, variant 3 | ||
| Orthology group | MCL10362 | Multiple-copy universal gene | ||
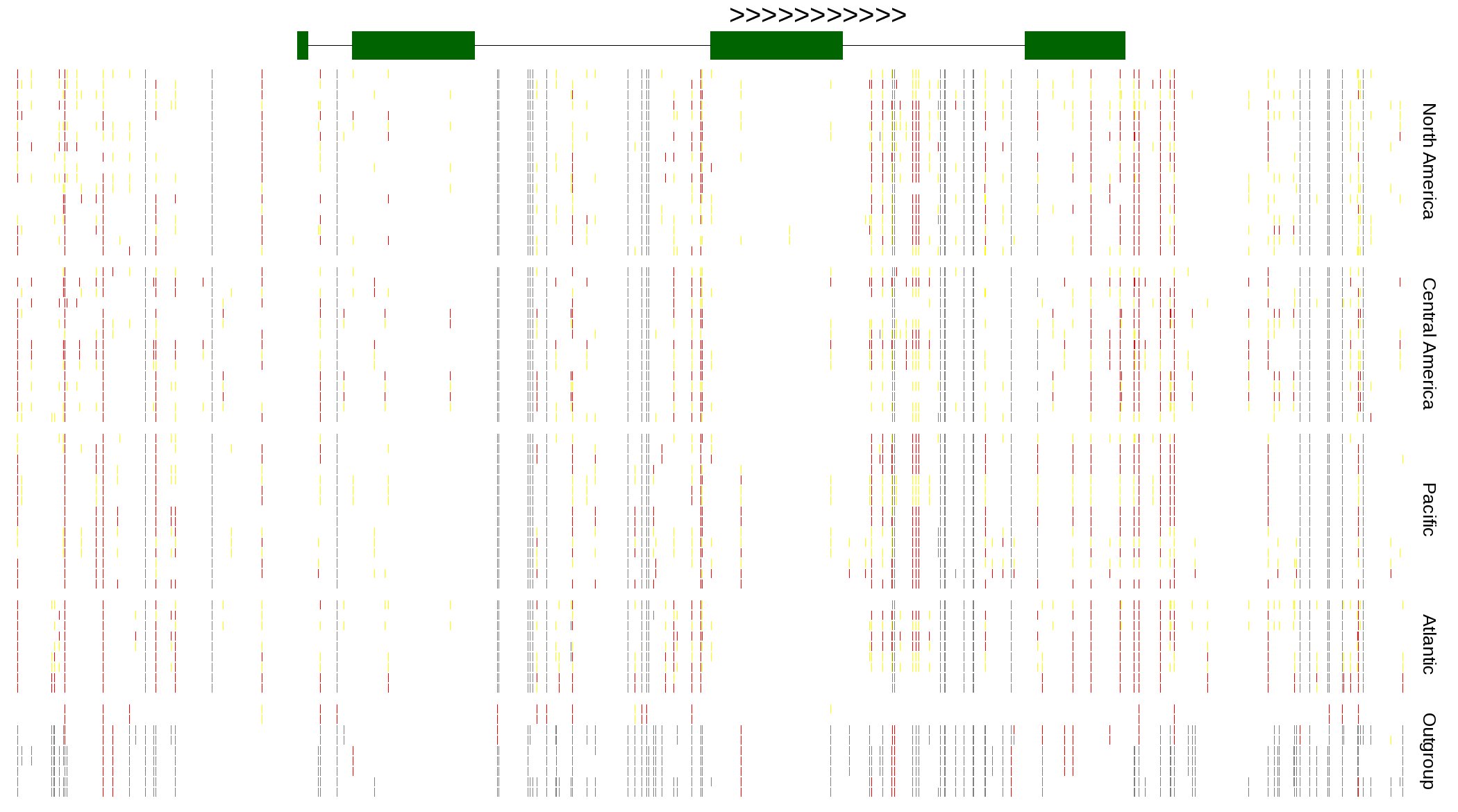
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS204688-TA
ATGGATGGACTTATTTTGAATACTGAACATCTATATACAGTTGCATTTCAAAACATTGTATCTCGTTATGGAAAAAATTATACATTTGAATTAAAAATGAGGCTAATGGGTTCGCAGTCCCACGAATTAGCGAAAATTATCACAGAAGAACTCGAATTACCCCTGACACCTGACGAATTTTTAGTTGAAACTAGAAAACAATTTCAAGAATTATTTCCACAGACAGAACTAATGCCAGGTGCTGAACGACTGATAAGACATTTGGACAATAAATGCATACCTATTGGGCTAGCGACAAGTTCCAGCGAGGATAGTTACCATCTCAAAGTAGATAAACACCATCAGGAGTTATTTTCTCTGTTCCCATACAAAACTTTTGGTTCTTCGGATCCTGATGTTGCAAGAGGAAAACCTTACCCTGATATATTTTTGGTTGCTGCTTCCAAATTTCCGGAAAATCCAAAAGTAGAACAGTGCCTTGTATTTGAGGATTCGGTAAATGGAGTGAGGGCAGGATTAGCAGCGGGGATGCAAGTTGTCATGGTGCCGGACCCTAGAGTCAACAAAATTCTCACCGAAGAAGCAACATTGGTATTAGGAAGTCTTGAAGAATTTAAACCAGAATTATTTGGGTTACCCCCATTTGAAGATTAA
>DPOGS204688-PA
MDGLILNTEHLYTVAFQNIVSRYGKNYTFELKMRLMGSQSHELAKIITEELELPLTPDEFLVETRKQFQELFPQTELMPGAERLIRHLDNKCIPIGLATSSSEDSYHLKVDKHHQELFSLFPYKTFGSSDPDVARGKPYPDIFLVAASKFPENPKVEQCLVFEDSVNGVRAGLAAGMQVVMVPDPRVNKILTEEATLVLGSLEEFKPELFGLPPFED-