| DPOGS204706 | ||
|---|---|---|
| Transcript | DPOGS204706-TA | 621 bp |
| Protein | DPOGS204706-PA | 206 aa |
| Genomic position | DPSCF300170 + 616109-617930 | |
| RNAseq coverage | 28x (Rank: top 76%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL008363 | 2e-49 | 50.52% | |
| Bombyx | BGIBMGA007484-TA | 1e-50 | 57.86% | |
| Drosophila | beta4GalNAcTA-PA | 2e-45 | 48.85% | |
| EBI UniRef50 | UniRef50_Q6J4T9 | 5e-53 | 50.77% | Beta 1,4-N-acetylgalactosaminyltransferase n=6 Tax=Arthropoda RepID=Q6J4T9_TRINI |
| NCBI RefSeq | XP_318033.4 | 2e-48 | 51.56% | AGAP004781-PA [Anopheles gambiae str. PEST] |
| NCBI nr blastp | gi|47156063 | 2e-52 | 50.77% | beta 1,4-N-acetylgalactosaminyltransferase [Trichoplusia ni] |
| NCBI nr blastx | gi|47156063 | 3e-51 | 50.77% | beta 1,4-N-acetylgalactosaminyltransferase [Trichoplusia ni] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0016757 | 8.8e-78 | transferase activity, transferring glycosyl groups | |
| GO:0005975 | 8.8e-78 | carbohydrate metabolic process | ||
| KEGG pathway | cel:Y73E7A.7 | 1e-42 | ||
| K07968 (B4GALT3) | maps-> | Glycosphingolipid biosynthesis - lacto and neolacto series | ||
| Glycosaminoglycan biosynthesis - keratan sulfate | ||||
| N-Glycan biosynthesis | ||||
| InterPro domain | [1-206] IPR003859 | 8.8e-78 | Galactosyltransferase, metazoa | |
| Orthology group | MCL30504 | Lepidoptera specific | ||
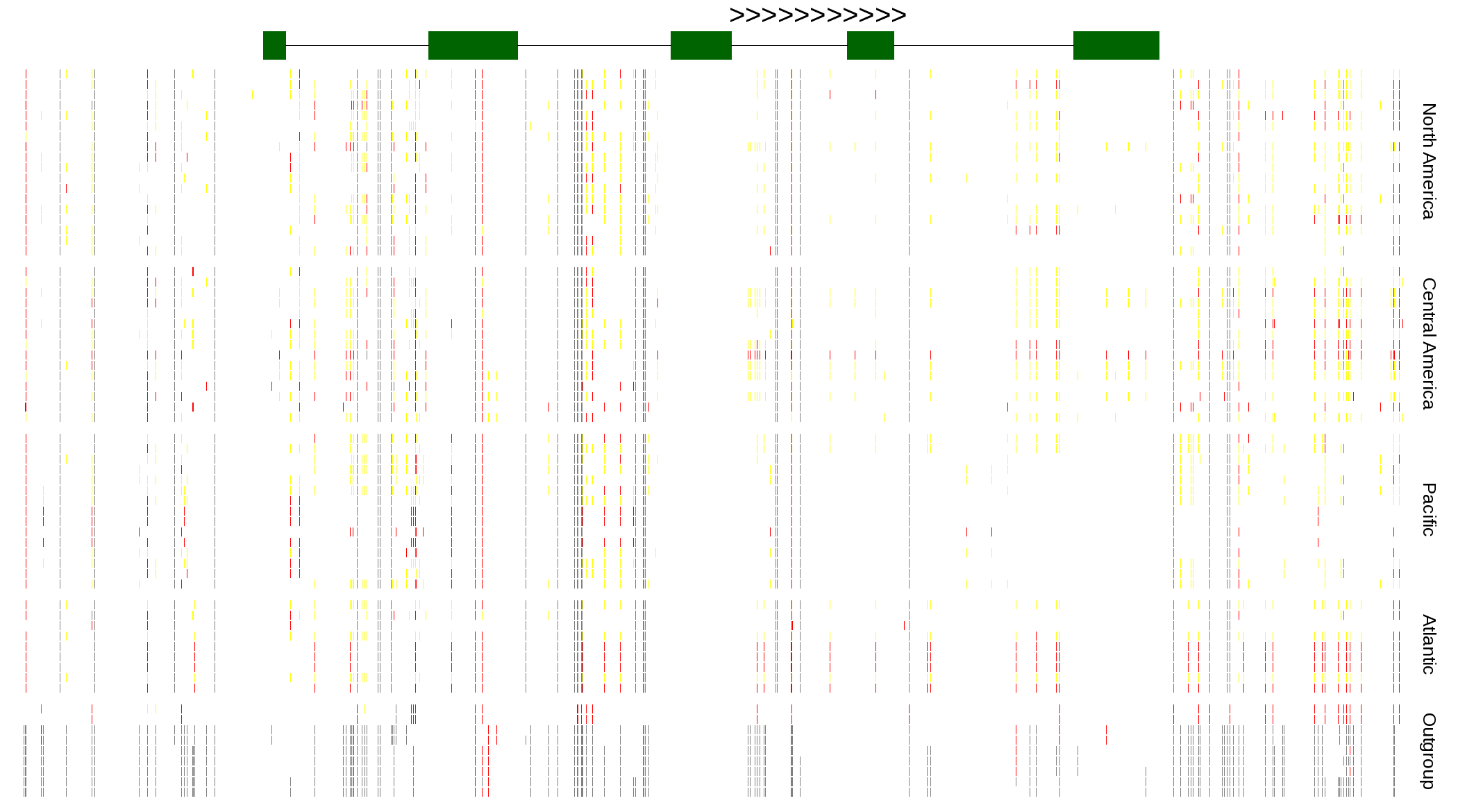
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS204706-TA
ATGAAACAGAAGTTGGAGTACCGGATATTCATTGTTGAACAGAAAGGTACAGATTTCTTCAATAGAGGGCGCCTCTTCAATGCTGGTTATTTGGAGGTACGGAAGTTTGGCAATTGGAAATGTGTAGTGTTCCACGACGTAGACCTGCTTCCTCTGGATGATAGAATACTCTACTCCTGTCCAATGTGGCCAAGACACATGTGTGGCACAGTCGTGGAGGTTAAGAATCCGAGTTTCCGAACACTATTCGGCGGAGTTTCCGCAATGATTCCACAGCATTTCGAGAAAGTTAACGGTTTCTCAAACGTATATTGGGGTTGGGGCGGCGAGGATAACGACTTATTTTGGAGGATTCGTGCCGTCGGTCTACCAATAGTTAGATACAACAAGCTTATAGCAAAATATACGTCACTTCAACATGACAAGTCGAAACCCAATACACTCAGATATAACCTTCTCAAAACTTTTGCGACACGTTTTTTACGAGACGGTCTTACAACTTTGGAATATGTCGTCGATAAAGTCACATTGCACCATCTCTACACGCACTTGATGCTGGATATAAACCCCAAGAAGAAAAACATTACAAAGATAATGTTAGAAGCATCTAGATGGATATAA
>DPOGS204706-PA
MKQKLEYRIFIVEQKGTDFFNRGRLFNAGYLEVRKFGNWKCVVFHDVDLLPLDDRILYSCPMWPRHMCGTVVEVKNPSFRTLFGGVSAMIPQHFEKVNGFSNVYWGWGGEDNDLFWRIRAVGLPIVRYNKLIAKYTSLQHDKSKPNTLRYNLLKTFATRFLRDGLTTLEYVVDKVTLHHLYTHLMLDINPKKKNITKIMLEASRWI-