| DPOGS204967 | ||
|---|---|---|
| Transcript | DPOGS204967-TA | 354 bp |
| Protein | DPOGS204967-PA | 117 aa |
| Genomic position | DPSCF301478 - 162-1315 | |
| RNAseq coverage | 40x (Rank: top 73%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL013573 | 2e-51 | 89.11% | |
| Bombyx | BGIBMGA006519-TA | 6e-49 | 76.85% | |
| Drosophila | Pxd-PA | 1e-30 | 54.13% | |
| EBI UniRef50 | UniRef50_Q01603 | 2e-28 | 54.13% | Peroxidase n=22 Tax=Neoptera RepID=PERO_DROME |
| NCBI RefSeq | XP_967241.1 | 2e-30 | 59.63% | PREDICTED: similar to AGAP010734-PA [Tribolium castaneum] |
| NCBI nr blastp | gi|91078176 | 4e-29 | 59.63% | PREDICTED: similar to AGAP010734-PA [Tribolium castaneum] |
| NCBI nr blastx | gi|91078176 | 2e-29 | 59.63% | PREDICTED: similar to AGAP010734-PA [Tribolium castaneum] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0006979 | 4.8e-19 | response to oxidative stress | |
| GO:0020037 | 4.8e-19 | heme binding | ||
| GO:0004601 | 4.8e-19 | peroxidase activity | ||
| GO:0055114 | 4.8e-19 | oxidation-reduction process | ||
| KEGG pathway | bta:525013 | 6e-07 | ||
| K00431 (TPO) | maps-> | Cytokine-cytokine receptor interaction | ||
| Autoimmune thyroid disease | ||||
| Tyrosine metabolism | ||||
| Hematopoietic cell lineage | ||||
| Jak-STAT signaling pathway | ||||
| InterPro domain | [18-117] IPR002007 | 4.8e-19 | Haem peroxidase, animal | |
| [24-117] IPR010255 | 2.3e-13 | Haem peroxidase | ||
| Orthology group | ||||
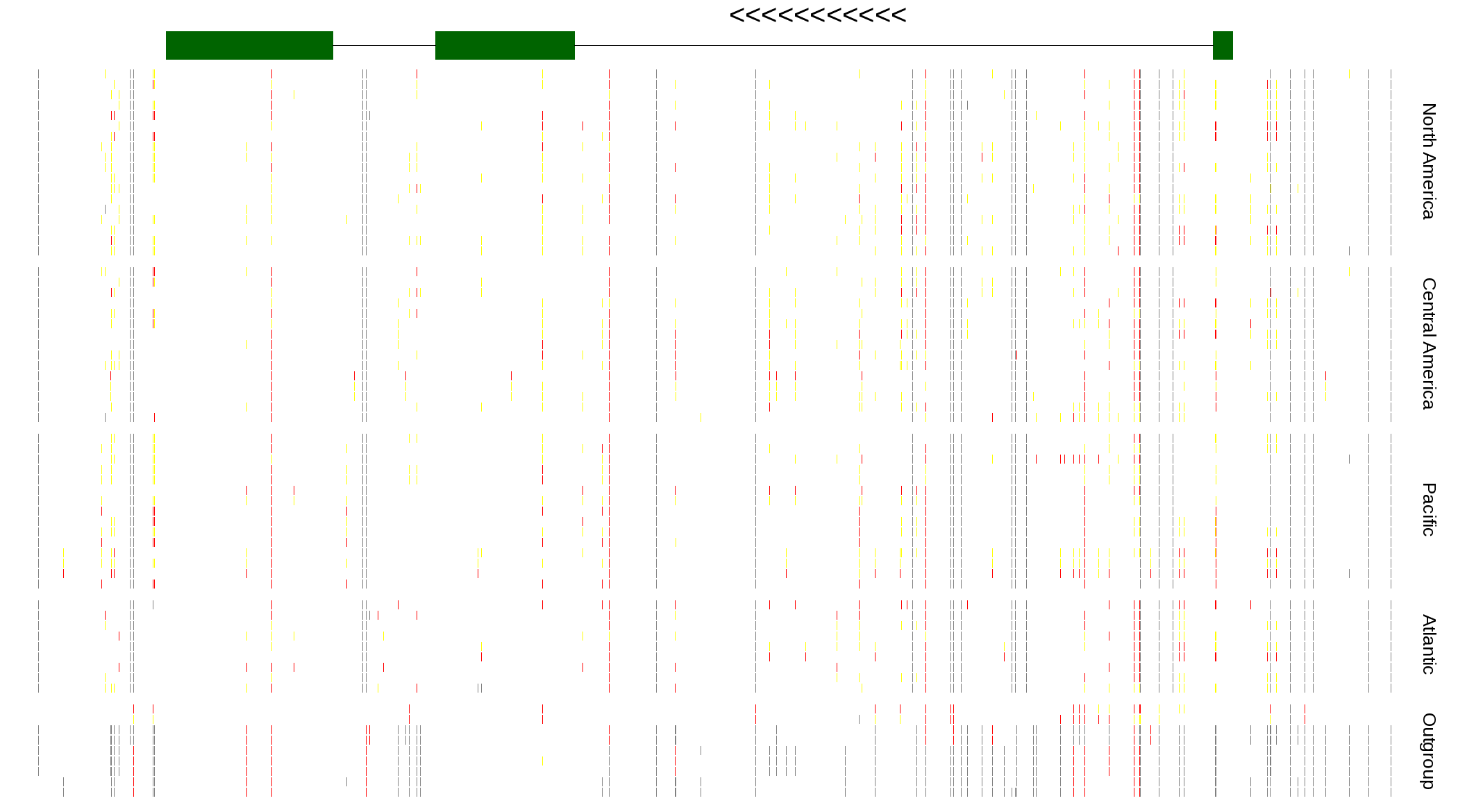
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS204967-TA
ATGATCTTCAGCATCATCGGAAGTGTTGAAAATATGATAAAGAACAAACTTATATATCCTGGCGCGAGGGGATACATCAATGATTATAACCCAAACGTCGATCCTACCGTGCTAGATGAACACGCCACCGCTGCGTTTAGACACTTCCATACGTTAATCCGCGGATATTTGCAATTGGTAACAGAAGACAGACACTTAGCTGGAATTGTTCGTCTGAGCGATTGGTTTAACCGGCCGCTTTTATTAGAAATCGAAAATGCTTTTGACGATTTAACAAGAGGACTTACTTACCAGCCTCAAGGATTTAGTGATCGATTTTTTGATAGCGAAATCACACAATACTTATTCAAGTAG
>DPOGS204967-PA
MIFSIIGSVENMIKNKLIYPGARGYINDYNPNVDPTVLDEHATAAFRHFHTLIRGYLQLVTEDRHLAGIVRLSDWFNRPLLLEIENAFDDLTRGLTYQPQGFSDRFFDSEITQYLFK-