| DPOGS205236 | ||
|---|---|---|
| Transcript | DPOGS205236-TA | 558 bp |
| Protein | DPOGS205236-PA | 185 aa |
| Genomic position | DPSCF300265 + 255004-265785 | |
| RNAseq coverage | 1x (Rank: top 93%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL014752 | 5e-28 | 70.00% | |
| Bombyx | BGIBMGA003874-TA | 2e-13 | 41.67% | |
| Drosophila | CG7786-PA | 9e-20 | 53.12% | |
| EBI UniRef50 | UniRef50_D6WCF2 | 1e-24 | 42.08% | Putative uncharacterized protein n=1 Tax=Tribolium castaneum RepID=D6WCF2_TRICA |
| NCBI RefSeq | XP_001600732.1 | 1e-22 | 36.36% | PREDICTED: similar to GA20586-PA [Nasonia vitripennis] |
| NCBI nr blastp | gi|270002434 | 4e-24 | 42.08% | hypothetical protein TcasGA2_TC004495 [Tribolium castaneum] |
| NCBI nr blastx | gi|270002434 | 1e-24 | 42.35% | hypothetical protein TcasGA2_TC004495 [Tribolium castaneum] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0006355 | 4.5e-07 | regulation of transcription, DNA-dependent | |
| GO:0043565 | 4.5e-07 | sequence-specific DNA binding | ||
| GO:0003700 | 4.5e-07 | sequence-specific DNA binding transcription factor activity | ||
| GO:0046983 | 4.5e-07 | protein dimerization activity | ||
| KEGG pathway | mdo:100012192 | 4e-12 | ||
| K09057 (HLF) | maps-> | Circadian rhythm - fly | ||
| InterPro domain | [93-121] IPR011700 | 4.5e-07 | Basic leucine zipper | |
| Orthology group | MCL17454 | Insect specific | ||
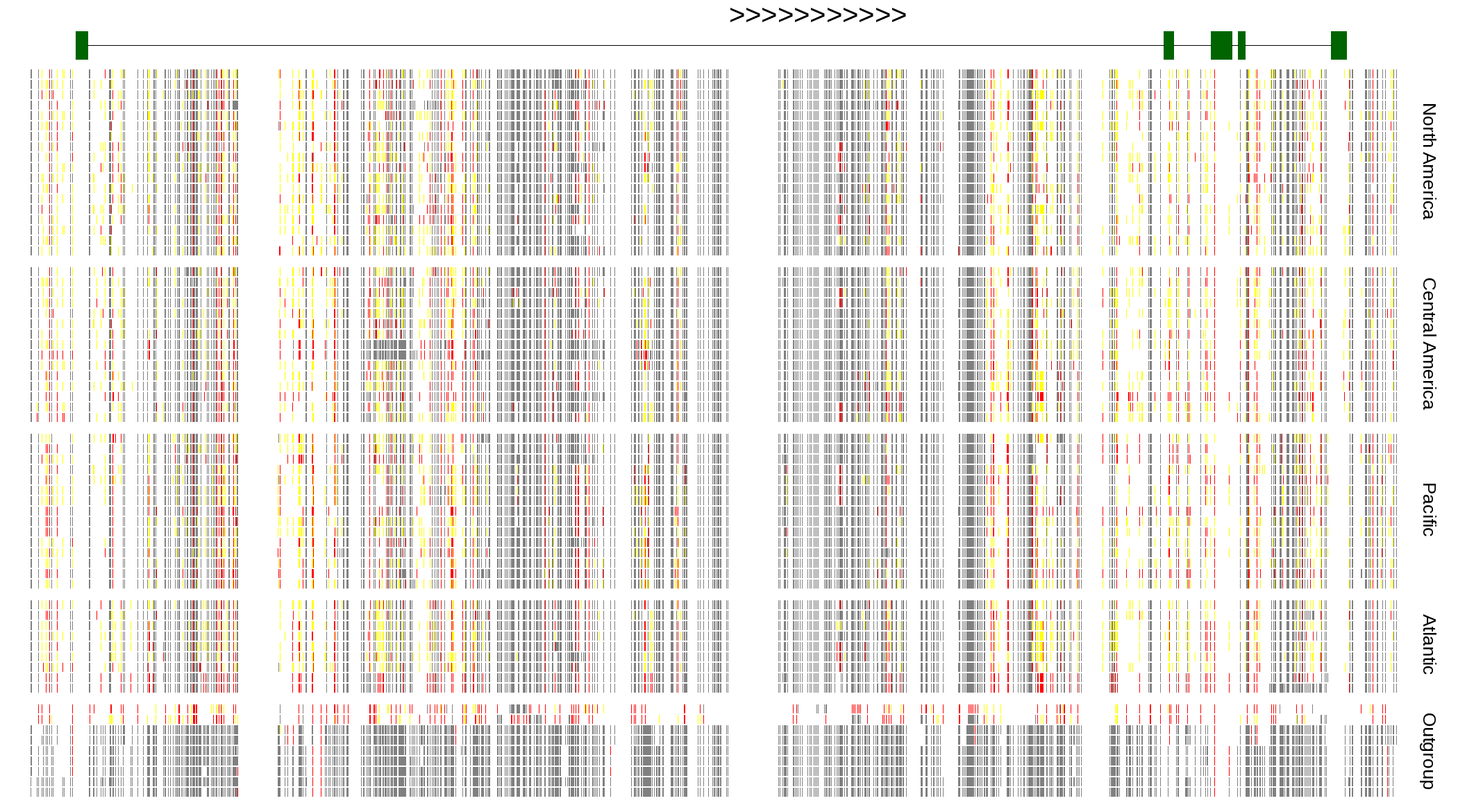
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS205236-TA
ATGGAAAACATGTACGGCTTCGATTCTTCATGCAGTCCGCACAGCGCTCTCACCGCGTTGAGACTTCTACGGAGCTACAACCACTTCGTAGTGCCGCCTGAAGCTCGTGGGATGGTACCACTCCTGACGGCTCTGCCACCACCGTGTTTACCAGCATATTGTCACCACCTACCACTGTCTTCACCACCTTTTGGGAACGAATCCACGAATAGCGTCACCTCGTTGCCAGAAGACGGGAGACGTCGCGGTGAGAAAAGACCCATACCCAGCGAGCTGAAAGACGAGAAGTACTTCGAGAGACGAAGACGGAATAATCAGGCGGCTAAGAAGAGCAGAGACGCCAGGAGAATGAGAGAGGATCAGTTCTATCCTGTCTTTATCTCCTTAGACCTCGGTTTGGACACAAAGTCCAAACAGAAACGTATTGCATGGAGAGCTTGCGTGTTGGAGCAAGAGAACGCGTCACTCCGCGCCCACGTGGCAGCGCTCACGCAAGAAACACGGGCGCTAAGAGCACTGCTGGCCAGAGACGAAGCGCCCACATCGACCACAGACTAG
>DPOGS205236-PA
MENMYGFDSSCSPHSALTALRLLRSYNHFVVPPEARGMVPLLTALPPPCLPAYCHHLPLSSPPFGNESTNSVTSLPEDGRRRGEKRPIPSELKDEKYFERRRRNNQAAKKSRDARRMREDQFYPVFISLDLGLDTKSKQKRIAWRACVLEQENASLRAHVAALTQETRALRALLARDEAPTSTTD-