| DPOGS205386 | ||
|---|---|---|
| Transcript | DPOGS205386-TA | 366 bp |
| Protein | DPOGS205386-PA | 121 aa |
| Genomic position | DPSCF300407 - 409182-413228 | |
| RNAseq coverage | 383x (Rank: top 31%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | % | |||
| Bombyx | BGIBMGA007425-TA | 3e-31 | 52.07% | |
| Drosophila | Pgm-PA | 3e-25 | 44.92% | |
| EBI UniRef50 | UniRef50_P00949 | 1e-23 | 44.63% | Phosphoglucomutase-1 n=21 Tax=cellular organisms RepID=PGM1_RABIT |
| NCBI RefSeq | XP_001608147.1 | 3e-26 | 46.28% | PREDICTED: similar to GA18703-PA [Nasonia vitripennis] |
| NCBI nr blastp | gi|262530078 | 8e-27 | 48.76% | phosphoglucomutase [Spodoptera exigua] |
| NCBI nr blastx | gi|262530078 | 4e-26 | 48.76% | phosphoglucomutase [Spodoptera exigua] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0005975 | 9.1e-10 | carbohydrate metabolic process | |
| GO:0016868 | 9.1e-10 | intramolecular transferase activity, phosphotransferases | ||
| KEGG pathway | tel:tlr1976 | 6e-27 | ||
| K01835 (E5.4.2.2, pgm) | maps-> | Streptomycin biosynthesis | ||
| Starch and sucrose metabolism | ||||
| Purine metabolism | ||||
| Galactose metabolism | ||||
| Pentose phosphate pathway | ||||
| Glycolysis / Gluconeogenesis | ||||
| Amino sugar and nucleotide sugar metabolism | ||||
| InterPro domain | [30-81] IPR005843 | 9.1e-10 | Alpha-D-phosphohexomutase, C-terminal | |
| Orthology group | ||||
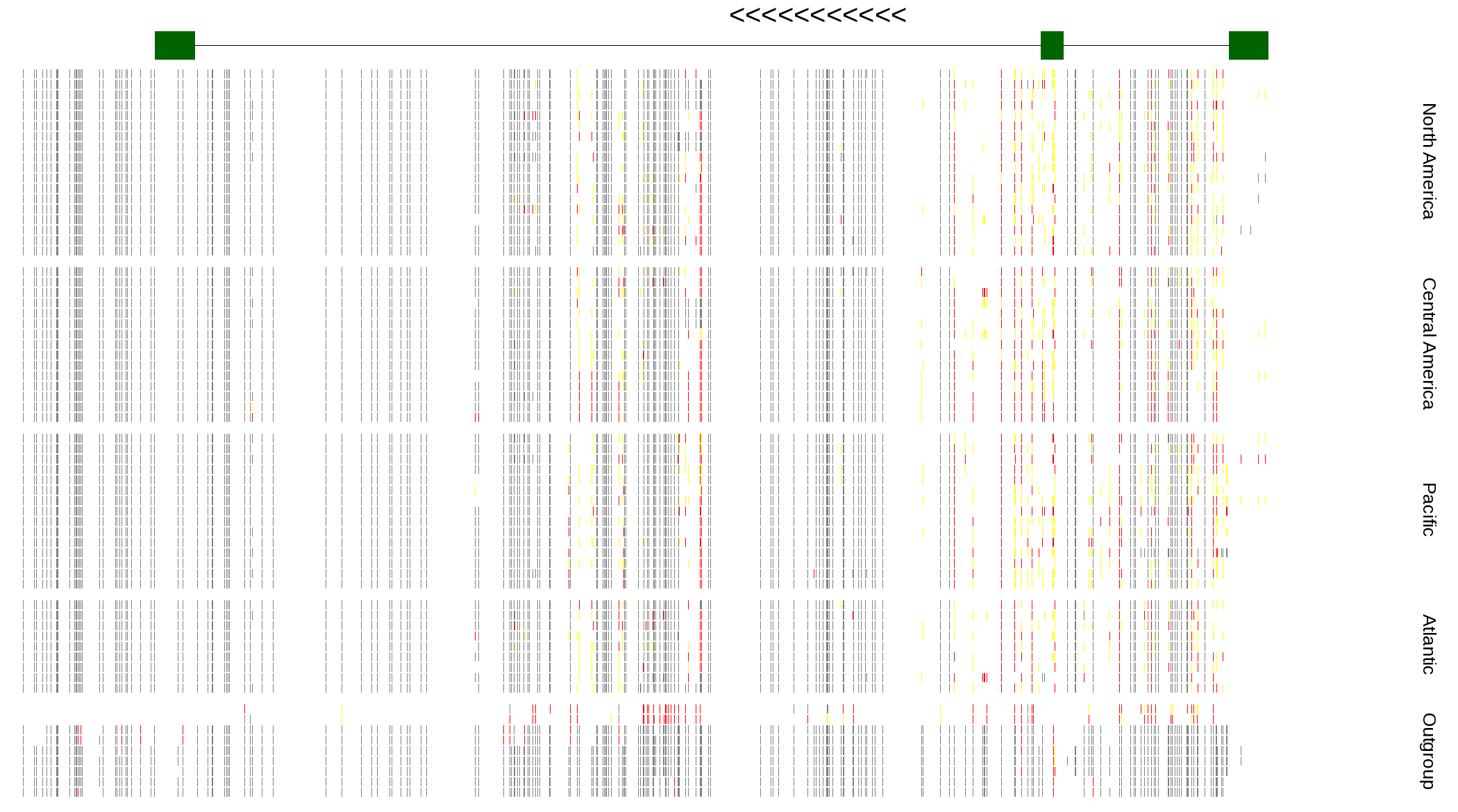
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS205386-TA
ATGACCGACCTGGAGCGGACGGTCACCGCCGAGGGGTTCGTGGGATCCAAGCATTATATACAGGATGTAGAGAACGCGGTCGTCGCCGCCGACAACTTCTCATACATGGACCCCATAGACCGCAGCGTCGCCTTGAACCAGGGTATTCGTCTGGTTTTTGAGAACGGCAGTCGCATCATATACAGGCTGAGCGGCACCGGCAGCCAGGGGGCCACGTTACGTATATATTTGGAAGCGTTCACTCCCTGTGACGGCGACATCCACCTGTCCAGCGAGGAGCGGCTTCAGTCCCTGGTGACGCTCGCCATGAGGTTCGGCAGAGTGAGGAGATACACCGGCAGGGACAAACCGACAGTCATCACGTAG
>DPOGS205386-PA
MTDLERTVTAEGFVGSKHYIQDVENAVVAADNFSYMDPIDRSVALNQGIRLVFENGSRIIYRLSGTGSQGATLRIYLEAFTPCDGDIHLSSEERLQSLVTLAMRFGRVRRYTGRDKPTVIT-