| DPOGS205897 | ||
|---|---|---|
| Transcript | DPOGS205897-TA | 654 bp |
| Protein | DPOGS205897-PA | 217 aa |
| Genomic position | DPSCF300089 - 379605-380326 | |
| RNAseq coverage | 445x (Rank: top 28%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL005510 | 3e-121 | 95.85% | |
| Bombyx | BGIBMGA007083-TA | 2e-86 | 99.33% | |
| Drosophila | CG32810-PB | 2e-76 | 71.72% | |
| EBI UniRef50 | UniRef50_Q9NXV2 | 1e-76 | 71.13% | BTB/POZ domain-containing protein KCTD5 n=39 Tax=Coelomata RepID=KCTD5_HUMAN |
| NCBI RefSeq | XP_002429337.1 | 1e-91 | 81.91% | BTB/POZ domain-containing protein KCTD5, putative [Pediculus humanus corporis] |
| NCBI nr blastp | gi|380029002 | 4e-95 | 81.22% | PREDICTED: BTB/POZ domain-containing protein KCTD5-like [Apis florea] |
| NCBI nr blastx | gi|380029002 | 3e-93 | 81.22% | PREDICTED: BTB/POZ domain-containing protein KCTD5-like [Apis florea] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0016020 | 3.3e-23 | membrane | |
| GO:0006813 | 3.3e-23 | potassium ion transport | ||
| GO:0005249 | 3.3e-23 | voltage-gated potassium channel activity | ||
| GO:0008076 | 3.3e-23 | voltage-gated potassium channel complex | ||
| GO:0005515 | 4.1e-19 | protein binding | ||
| KEGG pathway | ||||
| InterPro domain | [24-134] IPR011333 | 7.6e-31 | BTB/POZ fold | |
| [29-117] IPR003131 | 3.3e-23 | Potassium channel, voltage dependent, Kv, tetramerisation | ||
| [27-129] IPR000210 | 4.1e-19 | BTB/POZ-like | ||
| Orthology group | MCL12998 | Single-copy universal gene | ||
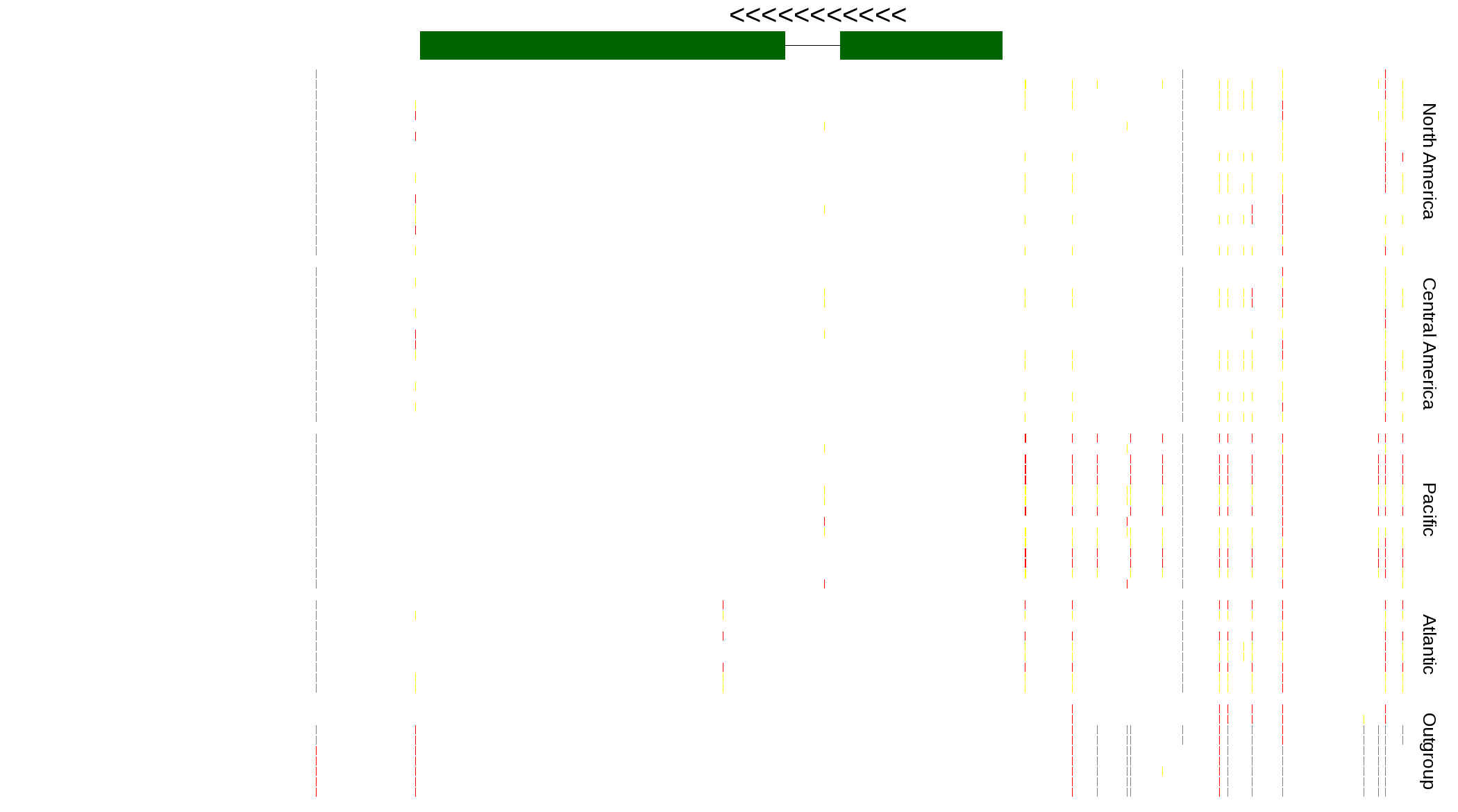
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS205897-TA
ATGGCGGGTTACGTAGGTGACGGTGATTTTTCTGAAGAAAATATACATAAATTTAACAATGAAAGAAGAAGTAGTAAGCAGTGGGTTAAACTAAATGTTGGGGGCACCTATTTTTTAACTACAAAAACCACGCTATCTAGGGACCCAAATTCATTTCTTAATCGATTAGTACAAGAGGACAGTGACTTAATTTCGGACAGAGACGAAACAGGCGCATATTTAATAGATAGGGATCCAACTTATTTTTCTCCAGTTCTGAATTATCTTCGACATGGAAAACTCGTTATTAATAATGATATAGCCGAGGAGGGAGTATTGGAGGAGGCAGAATTCTACAACATTACCGAACTTATCAGATTAGTCAAAGAGAGAATTTGTTTAAGGGAAAGAAGACCTCTCAAGGATTCCAAAAAACATGTGTACAGAGTCTTGCAATTTCATGAAGAGGAACTTACTCAAATGGTGTCTACTATGTCAGACGGTTGGAAGTTTGAACAGTTGATTAACATTGGTTCTCAGTATAACTATGGTAATGATGAACATGCTGAATTTCTCTGTGTTGTAAGTAGAGAATGTGGCAGCACCCTTAATAGTAATGACATAGAGCCCACTGATCGTGCAAAAGTATTACAACAGAAGGGCTCGAGAATGTGA
>DPOGS205897-PA
MAGYVGDGDFSEENIHKFNNERRSSKQWVKLNVGGTYFLTTKTTLSRDPNSFLNRLVQEDSDLISDRDETGAYLIDRDPTYFSPVLNYLRHGKLVINNDIAEEGVLEEAEFYNITELIRLVKERICLRERRPLKDSKKHVYRVLQFHEEELTQMVSTMSDGWKFEQLINIGSQYNYGNDEHAEFLCVVSRECGSTLNSNDIEPTDRAKVLQQKGSRM-