| DPOGS206141 | ||
|---|---|---|
| Transcript | DPOGS206141-TA | 765 bp |
| Protein | DPOGS206141-PA | 254 aa |
| Genomic position | DPSCF300028 + 1264710-1266034 | |
| RNAseq coverage | 200x (Rank: top 47%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL008776 | 6e-119 | 80.31% | |
| Bombyx | BGIBMGA000511-TA | 2e-117 | 77.95% | |
| Drosophila | scu-PA | 4e-94 | 64.82% | |
| EBI UniRef50 | UniRef50_Q99714 | 5e-89 | 65.48% | 3-hydroxyacyl-CoA dehydrogenase type-2 n=613 Tax=root RepID=HCD2_HUMAN |
| NCBI RefSeq | NP_001040424.1 | 6e-116 | 77.95% | 3-hydroxyacyl-CoA dehydrogenase [Bombyx mori] |
| NCBI nr blastp | gi|114051676 | 1e-114 | 77.95% | 3-hydroxyacyl-CoA dehydrogenase [Bombyx mori] |
| NCBI nr blastx | gi|114051676 | 9e-111 | 77.95% | 3-hydroxyacyl-CoA dehydrogenase [Bombyx mori] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0005488 | 1.5e-72 | binding | |
| GO:0008152 | 1.5e-30 | metabolic process | ||
| GO:0016491 | 1.5e-30 | oxidoreductase activity | ||
| KEGG pathway | dvi:Dvir_GJ16589 | 2e-93 | ||
| K08683 (HSD17B10) | maps-> | Alzheimer's disease | ||
| Valine, leucine and isoleucine degradation | ||||
| InterPro domain | [2-253] IPR016040 | 1.5e-72 | NAD(P)-binding domain | |
| [8-179] IPR002198 | 1.5e-30 | Short-chain dehydrogenase/reductase SDR | ||
| [6-23] IPR002347 | 2.8e-28 | Glucose/ribitol dehydrogenase | ||
| Orthology group | MCL13828 | Single-copy universal gene | ||
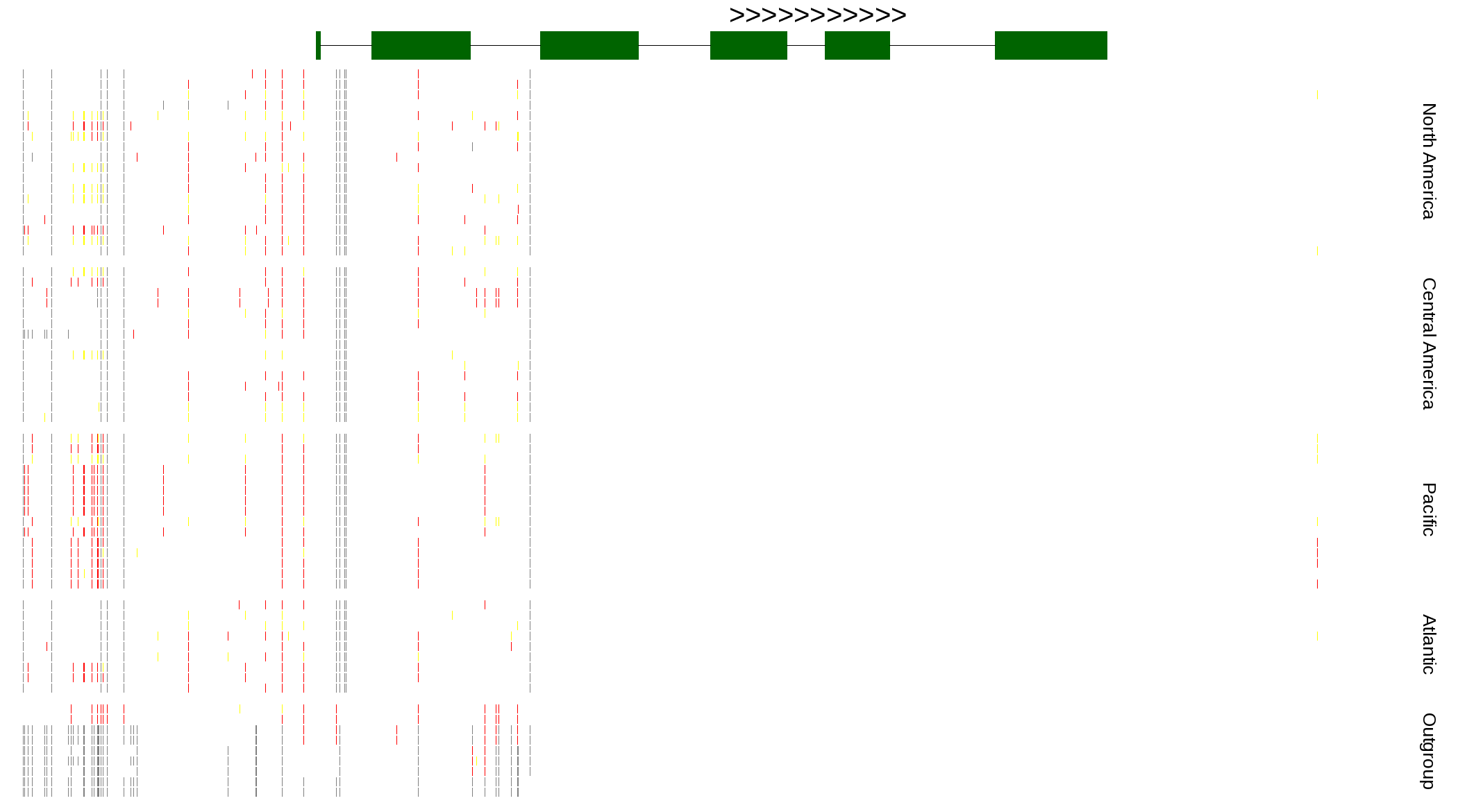
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS206141-TA
ATGTTTAAAGGTCTCGTAGGTTTAGTAACAGGAGGCGCCTCGGGACTCGGCAGGGGAACCGCTGAACATTTGGTAAAGCATGGAGGCAGGGTCGTTATTTGTGACCTCCCAAATAGTAAGGGCCGTGAGACAGCAAAACAGCTTGCAGGAAATGCTGCTTTTGTGCCAGTTGATGTTACATCAGAGGAGGATGTAAAAAATGCTCTACAGACAACGGTTGATAAGTTCGGGAGATTGGATGTGGTAGTCAACTGTGCAGGTATTATTACTGCTAGCAGAGTTTACAATGTCAAGAAAGATCAGCCGTTTGACCTAAAGGCTTTCCAAAGGACAATTGATGTGAATTTAATTGGAACATTCAATGTAATCCGTCTAGCAGTTGGTTTGATTTCTAAAAATGCTCCGGATGGGGACGGACAGCGGGGAGTTATTATTAATACTGCCAGTATCGCAGCTTTTGATGGACAAATAGGCCAAGCAGCTTATTCAGCGTCTAAGGCTGGTATTGTAGGGATGACGCTACCCATCGCTCGGGATCTAGCAAAACAAGGAATCAGAGTTGTAACCATCGCACCGGGTATGTTCCGGACACCACTAGTGGATGCAATGCCAGAAAAGGTTCTTCGTGAGTTGGAAGCAGGTGTACCTTTTCCCTCACGGTGCGGTCAGCCCCACGAGTTCGCTCTCTTGGTGCAAAGCATCATCACAAATCCCATGCTCAATGGCGAGACCATACGGATAGATGGCGCTTTAAGGATGCAGTAG
>DPOGS206141-PA
MFKGLVGLVTGGASGLGRGTAEHLVKHGGRVVICDLPNSKGRETAKQLAGNAAFVPVDVTSEEDVKNALQTTVDKFGRLDVVVNCAGIITASRVYNVKKDQPFDLKAFQRTIDVNLIGTFNVIRLAVGLISKNAPDGDGQRGVIINTASIAAFDGQIGQAAYSASKAGIVGMTLPIARDLAKQGIRVVTIAPGMFRTPLVDAMPEKVLRELEAGVPFPSRCGQPHEFALLVQSIITNPMLNGETIRIDGALRMQ-