| DPOGS206458 | ||
|---|---|---|
| Transcript | DPOGS206458-TA | 417 bp |
| Protein | DPOGS206458-PA | 138 aa |
| Genomic position | DPSCF300070 - 70372-71084 | |
| RNAseq coverage | 311x (Rank: top 36%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL012937 | 7e-76 | 99.28% | |
| Bombyx | BGIBMGA005469-TA | 3e-71 | 93.48% | |
| Drosophila | % | |||
| EBI UniRef50 | UniRef50_E2C245 | 5e-53 | 88.14% | Transcriptional adapter 2-alpha n=9 Tax=Coelomata RepID=E2C245_HARSA |
| NCBI RefSeq | XP_001599421.1 | 7e-63 | 87.88% | PREDICTED: similar to histone acetyltransferase, putative [Nasonia vitripennis] |
| NCBI nr blastp | gi|91082085 | 8e-60 | 82.61% | PREDICTED: similar to DNA-directed RNA polymerase II 16 kDa polypeptide [Tribolium castaneum] |
| NCBI nr blastx | gi|91082085 | 2e-58 | 82.61% | PREDICTED: similar to DNA-directed RNA polymerase II 16 kDa polypeptide [Tribolium castaneum] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0000166 | 6.6e-39 | nucleotide binding | |
| GO:0044237 | 6.6e-39 | cellular metabolic process | ||
| GO:0003824 | 6.6e-39 | catalytic activity | ||
| GO:0003899 | 3.7e-25 | DNA-directed RNA polymerase activity | ||
| GO:0006351 | 3.7e-25 | transcription, DNA-dependent | ||
| KEGG pathway | nvi:100115771 | 2e-62 | ||
| K03012 (RPB4) | maps-> | Huntington's disease | ||
| Purine metabolism | ||||
| Pyrimidine metabolism | ||||
| RNA polymerase | ||||
| InterPro domain | [20-137] IPR006590 | 7.2e-54 | RNA polymerase II, Rpb4, core | |
| [7-137] IPR010997 | 6.6e-39 | HRDC-like | ||
| [28-132] IPR005574 | 3.7e-25 | RNA polymerase II, Rpb4 | ||
| Orthology group | MCL17353 | Patchy | ||
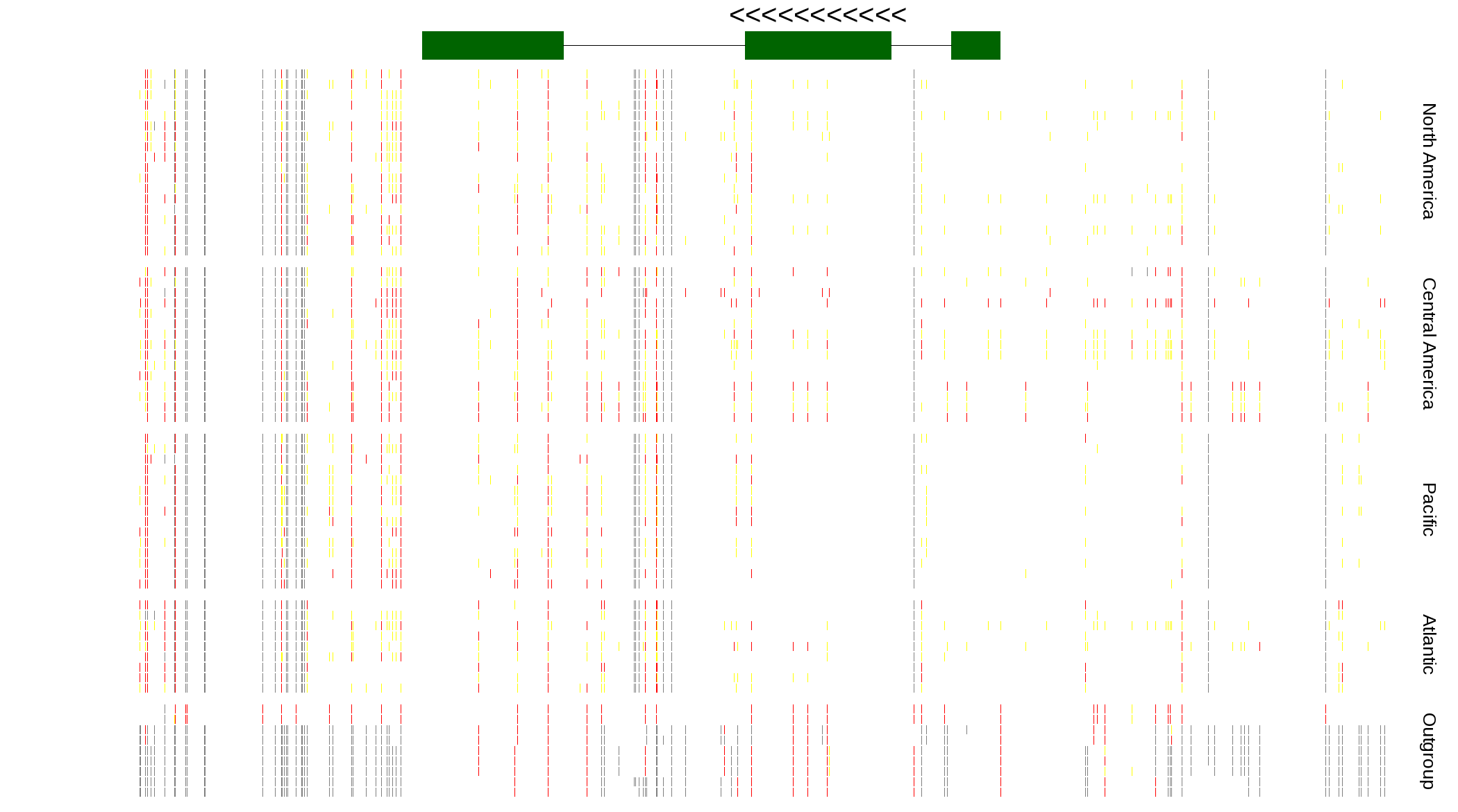
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS206458-TA
ATGTCTGGACCGCTACAAGACGTGGTAGAAGAAGACGCGGCGGATTTACAATTTCCAAAAGAATTCGAAAATGCAGAAACTTTATTAATATCCGAAGTCGATATGTTACTAGAGCATAGAAAAGCACAAAACGAATCCGCAGAGGAGGAACAAGAATTTTCTGAGGTTTTTATGAAAACACTCACCTATACAAACATGTTTAAAAAGTTTAAAAACAAAGAGACCATAACAGCTGTAAGAAACTTATTACAATCTAAAAAACTACATAAGTTTGAAGTAGCCAGTTTAGCAAATTTATGCCCCGAAACACCCGAAGAAGCTAAAGCTCTTATTCCATCACTTGAAGGGAGGTTTGAAGACGAGGAACTCAGAATATTGTTAGATGACATACAAACAAAACGAAGTATCCAATACTAA
>DPOGS206458-PA
MSGPLQDVVEEDAADLQFPKEFENAETLLISEVDMLLEHRKAQNESAEEEQEFSEVFMKTLTYTNMFKKFKNKETITAVRNLLQSKKLHKFEVASLANLCPETPEEAKALIPSLEGRFEDEELRILLDDIQTKRSIQY-