| DPOGS206909 | ||
|---|---|---|
| Transcript | DPOGS206909-TA | 612 bp |
| Protein | DPOGS206909-PA | 203 aa |
| Genomic position | DPSCF300001 - 1583706-1585178 | |
| RNAseq coverage | 83x (Rank: top 64%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL009426 | 4e-51 | 55.36% | |
| Bombyx | BGIBMGA007987-TA | 4e-30 | 35.62% | |
| Drosophila | PGRP-LB-PD | 9e-52 | 53.70% | |
| EBI UniRef50 | UniRef50_Q0KKW7 | 5e-60 | 57.59% | Peptidoglycan recognition protein B n=2 Tax=Bombycoidea RepID=Q0KKW7_SAMCR |
| NCBI RefSeq | XP_002069945.1 | 5e-52 | 54.19% | GK11791 [Drosophila willistoni] |
| NCBI nr blastp | gi|113208232 | 2e-59 | 57.59% | peptidoglycan recognition protein B [Samia cynthia ricini] |
| NCBI nr blastx | gi|113208232 | 3e-59 | 57.07% | peptidoglycan recognition protein B [Samia cynthia ricini] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0008745 | 2.4e-81 | N-acetylmuramoyl-L-alanine amidase activity | |
| GO:0009253 | 2.4e-81 | peptidoglycan catabolic process | ||
| GO:0008270 | 1.2e-58 | zinc ion binding | ||
| KEGG pathway | ||||
| InterPro domain | [1-199] IPR017331 | 2.6e-95 | Peptidoglycan recognition protein, PGRP-S | |
| [33-199] IPR015510 | 2.4e-81 | Peptidoglycan recognition protein | ||
| [26-197] IPR002502 | 7e-67 | N-acetylmuramoyl-L-alanine amidase domain | ||
| [32-175] IPR006619 | 1.2e-58 | Peptidoglycan recognition protein family domain, metazoa/bacteria | ||
| Orthology group | MCL17883 | Insect specific | ||
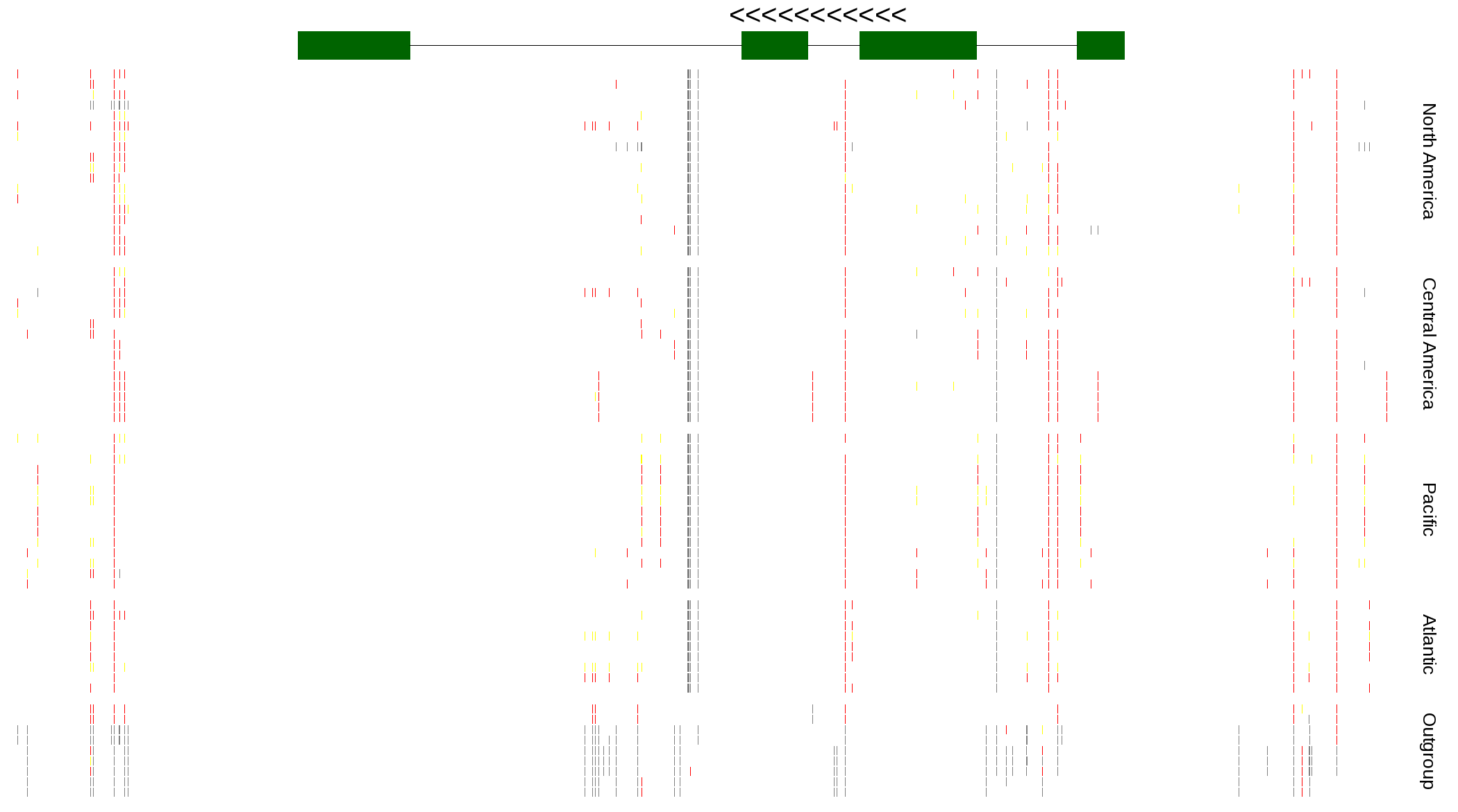
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS206909-TA
ATGACTCCTATTATTATCGGACTTTTTTGTGTGGCCGTCGGTGTAGTCGGATCTCCTTTACCACGTACACATTCAAGACACTATTCACATCAATTAGAATTTGTCTCGAAAGCGGACTGGGGAGGCAGGCACGCTACTTCGGTCACCAAACTAAAAACCCCCGTCCCTTTGGTGGTGATCCATCATAGTTACATCCCCGGCGCCTGTTATACGAAGGCAGCATGTTCTGCTGCAATGCAATCTATGCAAAACGCACACCAAATCACCAATGGCTGGGCTGATATTGGTTATAATTTCGCAGTTGGTAGTGATGGGAGAGTATACGAGGGGCGTGGTTTCAACCGCTTGGGTGCGCACGCGGTTGGTGTTAACGTAAAGAGCATTGGTATTGTCCTAATAGGAAATTGGGTCTCGGAAGTTCCTCCGAGAGTGCAACTGGAATCTGCGCAAGCATTGATTAAATTGGGAGTCGACCTCGGCTACATCAGTCCCGACTACAGCCTGATGGGCCACAGACAAGTCGGATCCACCGAGTGTCCCGGGGGCGCACTCCAAAAGGAAATATCCACTTGGCCCAAATTTCAACCAAATAGGGAATTCCACCTGACATGA
>DPOGS206909-PA
MTPIIIGLFCVAVGVVGSPLPRTHSRHYSHQLEFVSKADWGGRHATSVTKLKTPVPLVVIHHSYIPGACYTKAACSAAMQSMQNAHQITNGWADIGYNFAVGSDGRVYEGRGFNRLGAHAVGVNVKSIGIVLIGNWVSEVPPRVQLESAQALIKLGVDLGYISPDYSLMGHRQVGSTECPGGALQKEISTWPKFQPNREFHLT-