| DPOGS208404 | ||
|---|---|---|
| Transcript | DPOGS208404-TA | 723 bp |
| Protein | DPOGS208404-PA | 240 aa |
| Genomic position | DPSCF300241 - 2365-14885 | |
| RNAseq coverage | 1588x (Rank: top 8%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL013819 | 2e-73 | 91.02% | |
| Bombyx | BGIBMGA013112-TA | 2e-44 | 49.38% | |
| Drosophila | CG7656-PD | 1e-103 | 84.06% | |
| EBI UniRef50 | UniRef50_Q712K3 | 2e-87 | 75.65% | Ubiquitin-conjugating enzyme E2 R2 n=143 Tax=Metazoa RepID=UB2R2_HUMAN |
| NCBI RefSeq | XP_394314.2 | 1e-105 | 85.23% | PREDICTED: similar to CG7656-PC, isoform C [Apis mellifera] |
| NCBI nr blastp | gi|66504238 | 2e-104 | 85.23% | PREDICTED: ubiquitin-conjugating enzyme E2 R2-like [Apis mellifera] |
| NCBI nr blastx | gi|170030354 | 2e-118 | 85.36% | ubiquitin-conjugating enzyme E2r [Culex quinquefasciatus] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0016881 | 1.2e-48 | acid-amino acid ligase activity | |
| KEGG pathway | ame:410838 | 4e-105 | ||
| K02207 (UBE2R, UBC3, CDC34) | maps-> | Ubiquitin mediated proteolysis | ||
| InterPro domain | [3-203] IPR016135 | 1.6e-70 | Ubiquitin-conjugating enzyme/RWD-like | |
| [14-154] IPR000608 | 1.2e-48 | Ubiquitin-conjugating enzyme, E2 | ||
| Orthology group | MCL15677 | Multiple-copy universal gene | ||
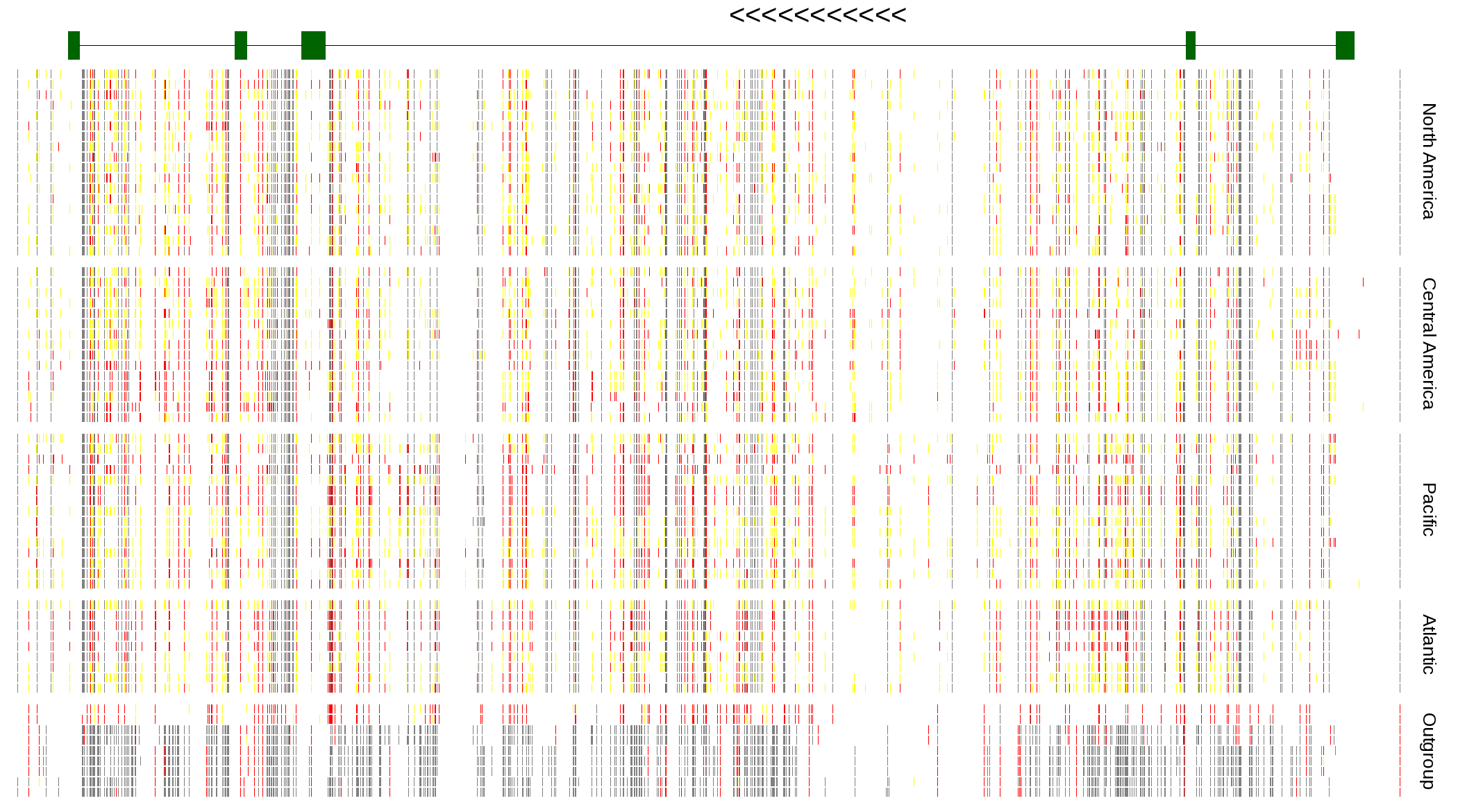
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS208404-TA
ATGTCTAGTACGGTACCGACGAGTTCTGCATTGAGGGCTTTGGCCTTAGAATATAAGAGCCTACAAGAAGAACCCGTGGAAGGGTTTAGAGTGAAATTATTGGGTGAGGACAATTTATTTGAATGGGAGGTTGCAATTTTCGGGCCACCAGACACGCTTTACCAAGGAGGATATTTCAAGGCTCATATGAAATTTCCACCGGACTATCCATATTCTCCACCATCTATAAAGTTCCTGACTAAAGTTTGGCATCCGAATGTTTATGAGAACGGCGATCTCTGCATATCAATTCTGCATCCCCCCGTGGACGACCCACAGTCTGGTGAGCTGCCCTGCGAGCGTTGGAACCCCACCCAGTCCGTGCGCACGGTGCTGTTGTCCGTGATCTCGCTATTGAACGAACCGAACACCTTCAGCCCGGCTAACGTGGACGCCAGCGTCATGTACCGCCGCTGGCGCGAGTCCAAGGGCAAGGATAAGGAGTACGAAAATATTATCAGGAAACAAGCCCAAGTGGCTCGGATGGAGGCTGAGAAGGAAGGTATAGTGGTGCCGCTCACTTTAGAAGATTATTGTATTAAGACACAGGTCAAGTCCTCGACACAGGAACCGCAGCTGGATATGACTGATTTCTATGACGACGATTACGATGATCTGGACACGGATTCCGAAGGCGAGCTTGGAGGTGAGGATGGAGACGACAGCGGCAACGGGGAGTCGTGA
>DPOGS208404-PA
MSSTVPTSSALRALALEYKSLQEEPVEGFRVKLLGEDNLFEWEVAIFGPPDTLYQGGYFKAHMKFPPDYPYSPPSIKFLTKVWHPNVYENGDLCISILHPPVDDPQSGELPCERWNPTQSVRTVLLSVISLLNEPNTFSPANVDASVMYRRWRESKGKDKEYENIIRKQAQVARMEAEKEGIVVPLTLEDYCIKTQVKSSTQEPQLDMTDFYDDDYDDLDTDSEGELGGEDGDDSGNGES-