| DPOGS209134 | ||
|---|---|---|
| Transcript | DPOGS209134-TA | 465 bp |
| Protein | DPOGS209134-PA | 154 aa |
| Genomic position | DPSCF300061 - 824894-827484 | |
| RNAseq coverage | 4871x (Rank: top 3%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL014780 | 2e-63 | 76.62% | |
| Bombyx | BGIBMGA001307-TA | 3e-74 | 85.71% | |
| Drosophila | Sod-PA | 6e-55 | 68.83% | |
| EBI UniRef50 | UniRef50_P28755 | 1e-57 | 73.38% | Superoxide dismutase [Cu-Zn] n=35 Tax=root RepID=SODC_CERCA |
| NCBI RefSeq | NP_001037084.1 | 6e-73 | 85.71% | superoxide dismutase [Cu-Zn] [Bombyx mori] |
| NCBI nr blastp | gi|112982998 | 1e-71 | 85.71% | superoxide dismutase [Cu-Zn] [Bombyx mori] |
| NCBI nr blastx | gi|112982998 | 2e-74 | 85.71% | superoxide dismutase [Cu-Zn] [Bombyx mori] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0004784 | 3.7e-89 | superoxide dismutase activity | |
| GO:0055114 | 3.7e-89 | oxidation-reduction process | ||
| GO:0006801 | 1e-60 | superoxide metabolic process | ||
| GO:0046872 | 1e-60 | metal ion binding | ||
| KEGG pathway | cqu:CpipJ_CPIJ016031 | 3e-59 | ||
| K04565 (E1.15.1.1C, sodC, SOD1) | maps-> | Huntington's disease | ||
| Peroxisome | ||||
| Amyotrophic lateral sclerosis (ALS) | ||||
| Prion diseases | ||||
| InterPro domain | [1-152] IPR024134 | 3.7e-89 | Superoxide dismutase (Cu/Zn) / chaperones | |
| [3-151] IPR001424 | 1e-60 | Superoxide dismutase, copper/zinc binding domain | ||
| Orthology group | MCL14401 | Single-copy universal gene | ||
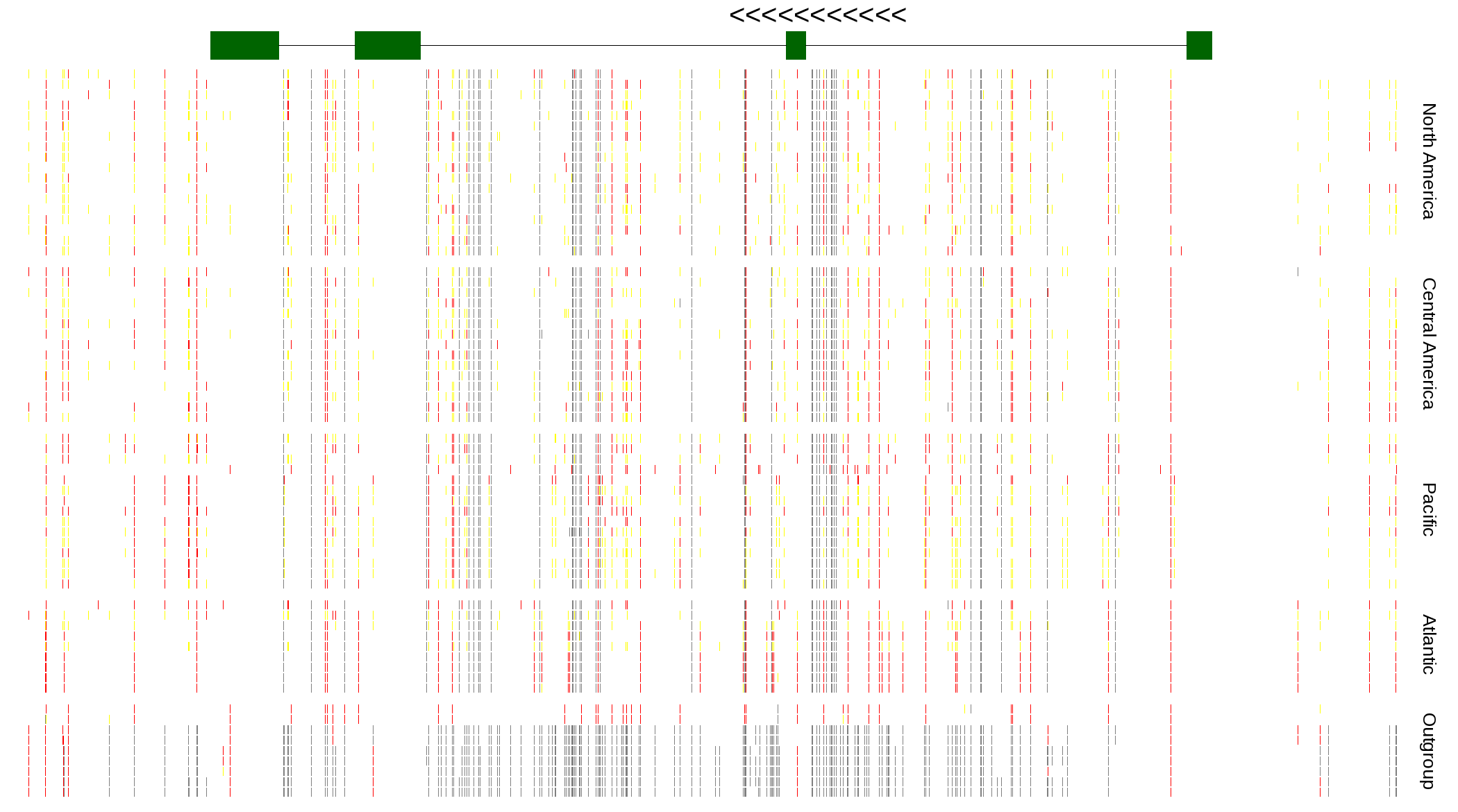
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS209134-TA
ATGCCGGCCAAAGCGGTTTGCGTTCTCAATGGAGATGTTAGCGGCACTGTTTTCTTCGATCAGAAGGACGACAAAGCCCCAGTTGTCGTGACGGGTGAAGTGAAAGGGCTTTCAAAGGGCAAGCACGGTTTCCACATCCATGAGTTTGGGGACAACACAAACGGCTGCACATCGGCCGGACCTCACTTCAATCCACAGAAGCAAGACCACGGTGCCCCTGATGCTGCTATCCGACATGTCGGAGACCTTGGAAATATTGAAGCCGGCAGTGACGGAGGTGTCACTAAGGTATGCATCCAAGATTCCCAGATCTCTCTGTGCGGCCCGAATAGCATCATTGGCCGTACTCTGGTTGTGCACGCTGATCCTGACGACCTCGGCATCGGCGGTCATGAACTGAGCAAGACGACTGGCAATGCCGGAGCTCGGGTCGCTTGTGGGGTTATTGGCCTCGCTAAGATCTAA
>DPOGS209134-PA
MPAKAVCVLNGDVSGTVFFDQKDDKAPVVVTGEVKGLSKGKHGFHIHEFGDNTNGCTSAGPHFNPQKQDHGAPDAAIRHVGDLGNIEAGSDGGVTKVCIQDSQISLCGPNSIIGRTLVVHADPDDLGIGGHELSKTTGNAGARVACGVIGLAKI-