| DPOGS209377 | ||
|---|---|---|
| Transcript | DPOGS209377-TA | 528 bp |
| Protein | DPOGS209377-PA | 175 aa |
| Genomic position | DPSCF300118 + 155241-155768 | |
| RNAseq coverage | 134x (Rank: top 56%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL021147 | 2e-72 | 75.93% | |
| Bombyx | BGIBMGA005699-TA | 6e-67 | 67.90% | |
| Drosophila | % | |||
| EBI UniRef50 | UniRef50_P53368 | 3e-42 | 49.68% | 7,8-dihydro-8-oxoguanine triphosphatase n=36 Tax=Coelomata RepID=8ODP_MOUSE |
| NCBI RefSeq | XP_394241.2 | 1e-42 | 49.07% | PREDICTED: similar to 7,8-dihydro-8-oxoguanine triphosphatase (8-oxo-dGTPase) (Nucleoside diphosphate-linked moiety X motif 1) (Nudix motif 1) [Apis mellifera] |
| NCBI nr blastp | gi|383863420 | 1e-43 | 53.16% | PREDICTED: 7,8-dihydro-8-oxoguanine triphosphatase-like [Megachile rotundata] |
| NCBI nr blastx | gi|383863420 | 2e-44 | 53.16% | PREDICTED: 7,8-dihydro-8-oxoguanine triphosphatase-like [Megachile rotundata] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0006281 | 7.1e-43 | DNA repair | |
| GO:0008413 | 7.1e-43 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity | ||
| GO:0016787 | 6.6e-33 | hydrolase activity | ||
| KEGG pathway | ||||
| InterPro domain | [4-21] IPR003563 | 7.1e-43 | 7,8-dihydro-8-oxoguanine triphosphatase | |
| [8-155] IPR000086 | 6.6e-33 | NUDIX hydrolase domain | ||
| [1-158] IPR015797 | 2.2e-24 | NUDIX hydrolase domain-like | ||
| Orthology group | MCL18307 | Patchy | ||
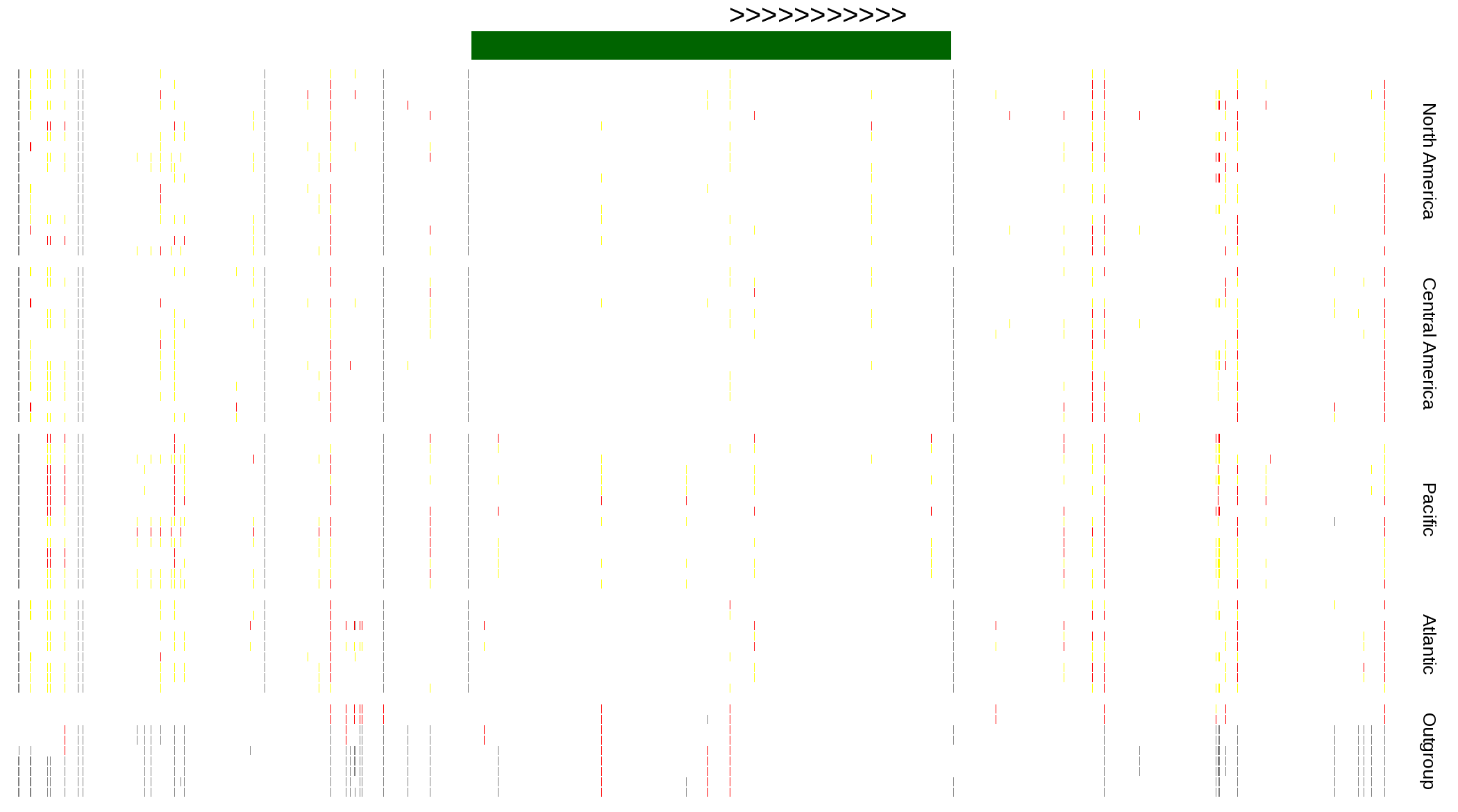
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS209377-TA
ATGGTCTTAAAGAAGATTTATACTCTTGTGTTTTTACGAAATGAAAAGAAGATTTTATTAGGATTGAAAAAACGTGGTTTTGGGATTAACAAATGGAACGGTTTTGGCGGCAAGGTGGAACAAAACGAATCTGTTATAGAGGCCGCGATAAGAGAAATGAAAGAAGAATGCTGTGTAACTGTTAAGAAAGATGATTTGAAGAGTGTCGGACAATTAGAATTTACTTTCGAAGGGGACCCAGTTATGATGGACGTTCGAGTGTTCTCAGGAGTTGTTTATTATGGGACCCCTACTGAAACTGAGGAAATGGCGCCGAAATGGTTTACGTACGACAGTATCCCTTTCGATGACATGTGGCCAGATGACAGACTGTGGTTCCCTTACATGTTAAATAACAAACTGTTCTATGGCAAATTTCATTACAAAGATTTAGACACAATATCGAACTATGATATTCGAGAACTCAAATCAATGGATTCATTCTACGCTTCTAGAGAACAGAATATTTCTCCGAAAGAAAAAAATTAA
>DPOGS209377-PA
MVLKKIYTLVFLRNEKKILLGLKKRGFGINKWNGFGGKVEQNESVIEAAIREMKEECCVTVKKDDLKSVGQLEFTFEGDPVMMDVRVFSGVVYYGTPTETEEMAPKWFTYDSIPFDDMWPDDRLWFPYMLNNKLFYGKFHYKDLDTISNYDIRELKSMDSFYASREQNISPKEKN-