| DPOGS210017 | ||
|---|---|---|
| Transcript | DPOGS210017-TA | 717 bp |
| Protein | DPOGS210017-PA | 238 aa |
| Genomic position | DPSCF300327 + 125211-131579 | |
| RNAseq coverage | 1x (Rank: top 94%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL007624 | 3e-36 | 44.76% | |
| Bombyx | BGIBMGA008409-TA | 2e-18 | 88.46% | |
| Drosophila | btn-PA | 6e-23 | 77.94% | |
| EBI UniRef50 | UniRef50_B0X7T8 | 2e-23 | 82.81% | Putative uncharacterized protein n=1 Tax=Culex quinquefasciatus RepID=B0X7T8_CULQU |
| NCBI RefSeq | XP_001865710.1 | 4e-24 | 82.81% | conserved hypothetical protein [Culex quinquefasciatus] |
| NCBI nr blastp | gi|170060229 | 7e-23 | 82.81% | conserved hypothetical protein [Culex quinquefasciatus] |
| NCBI nr blastx | gi|307204955 | 5e-22 | 85.48% | Homeobox protein MOX-2 [Harpegnathos saltator] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0003677 | 2.4e-25 | DNA binding | |
| GO:0006355 | 2.4e-25 | regulation of transcription, DNA-dependent | ||
| GO:0043565 | 6.7e-25 | sequence-specific DNA binding | ||
| GO:0003700 | 6.7e-25 | sequence-specific DNA binding transcription factor activity | ||
| GO:0005515 | 8.4e-22 | protein binding | ||
| KEGG pathway | ||||
| InterPro domain | [167-228] IPR012287 | 2.4e-25 | Homeodomain-related | |
| [169-231] IPR001356 | 6.7e-25 | Homeobox | ||
| [153-226] IPR009057 | 8.4e-22 | Homeodomain-like | ||
| [191-202] IPR020479 | 2.9e-07 | Homeobox, eukaryotic | ||
| Orthology group | MCL16978 | Patchy | ||
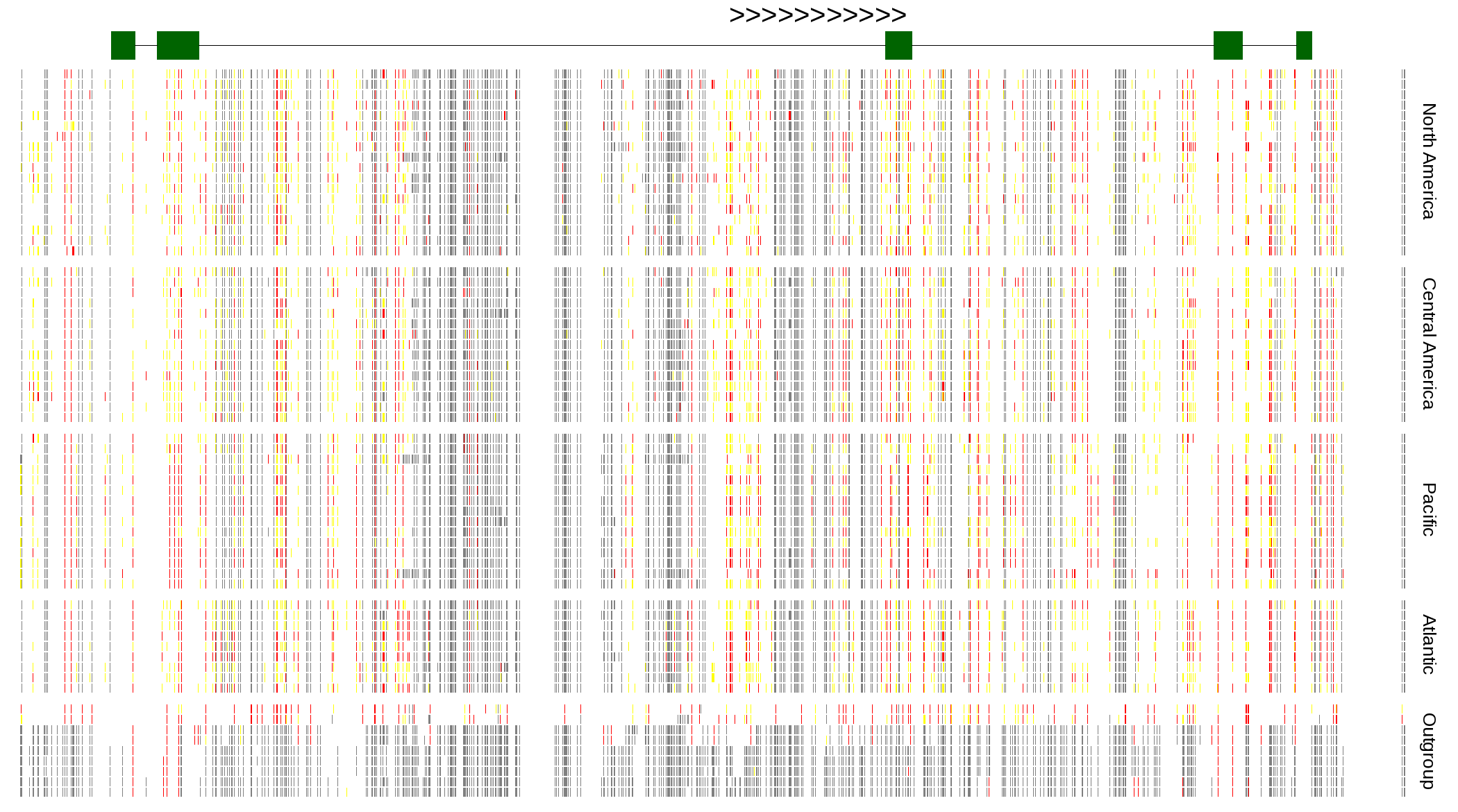
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS210017-TA
ATGATCGAACCAAACCCCGCCCAGGACCCGCCTTCAGGCGCGCGCGCTGATGAAACGAGTCATGAATTTCATGGAAGGAAGACGTGCCCAGTTGTTGCTTCTTATAATATAACAGTGGCGAAGTCTAAAATACATAATTTCGAAACGGTCCGCCAAGAAATACCTCAAAATACAGAGTTTCCGAATTATTTCCAAATGCTGTATGAGTACGGCGGCGAGAGGATCGAGCAGATGCAAGGACATCAACATCCTGGGGTTCAGGTGTTTGATTTTCAGAACTGGTGGAGCGAACAGGTTGTCAAAGAAGAGGACGAGGATCCGGGTGCTACGACTAGTCTTCAGGAAGGAGGTCATACATTTTTAATACAAGACCCAAAATCAAATTTGTTGGAGCCCATGAAGAACAAACTACACCTCAGGGCGGACACTTGCTTTTACCTGCAAAGCATGAGGCCATTCATGAATAGTGCCTACTGTTATAGCTCTGAACTGAGGTTGCCTGCGGCTCGCAAAGAGAGGACAGCGTTCACAAAGACACAGATCAAGAGCCTGGAGGCTGAGTTCGAGCGAGCCAACTATCTCACGCGACTTAGAAGATACGAGATTGCGGTCGCCTTACATCTCACAGAGAGACAGGTGAAGGTGTGGTTTCAGAACAGAAGAATGAAATGGAAGCGAACCAAAGGCGTCGCGAAGAGCGCCGGGAGACTCGCCTAG
>DPOGS210017-PA
MIEPNPAQDPPSGARADETSHEFHGRKTCPVVASYNITVAKSKIHNFETVRQEIPQNTEFPNYFQMLYEYGGERIEQMQGHQHPGVQVFDFQNWWSEQVVKEEDEDPGATTSLQEGGHTFLIQDPKSNLLEPMKNKLHLRADTCFYLQSMRPFMNSAYCYSSELRLPAARKERTAFTKTQIKSLEAEFERANYLTRLRRYEIAVALHLTERQVKVWFQNRRMKWKRTKGVAKSAGRLA-