| DPOGS210094 | ||
|---|---|---|
| Transcript | DPOGS210094-TA | 633 bp |
| Protein | DPOGS210094-PA | 210 aa |
| Genomic position | DPSCF300017 + 590963-592570 | |
| RNAseq coverage | 2077x (Rank: top 6%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL015471 | 1e-84 | 90.00% | |
| Bombyx | BGIBMGA012674-TA | 9e-103 | 85.57% | |
| Drosophila | eIF-4E-PE | 1e-64 | 55.45% | |
| EBI UniRef50 | UniRef50_P48598 | 1e-62 | 55.45% | Eukaryotic translation initiation factor 4E n=31 Tax=Eumetazoa RepID=IF4E_DROME |
| NCBI RefSeq | NP_001040379.1 | 2e-106 | 86.19% | mRNA cap-binding protein eIF4E [Bombyx mori] |
| NCBI nr blastp | gi|15290529 | 2e-110 | 90.48% | mRNA cap-binding protein eIF4E [Spodoptera frugiperda] |
| NCBI nr blastx | gi|15290529 | 1e-110 | 90.48% | mRNA cap-binding protein eIF4E [Spodoptera frugiperda] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0006413 | 3.4e-95 | translational initiation | |
| GO:0003743 | 3.4e-95 | translation initiation factor activity | ||
| GO:0003723 | 3.4e-95 | RNA binding | ||
| GO:0005737 | 3.4e-95 | cytoplasm | ||
| KEGG pathway | tca:662295 | 2e-71 | ||
| K03259 (eIF-4E, EIF4E) | maps-> | Insulin signaling pathway | ||
| mTOR signaling pathway | ||||
| InterPro domain | [12-210] IPR001040 | 3.4e-95 | Translation Initiation factor eIF- 4e | |
| [6-205] IPR023398 | 6.1e-72 | Translation Initiation factor eIF- 4e-like domain | ||
| Orthology group | MCL12173 | Multiple-copy universal gene | ||
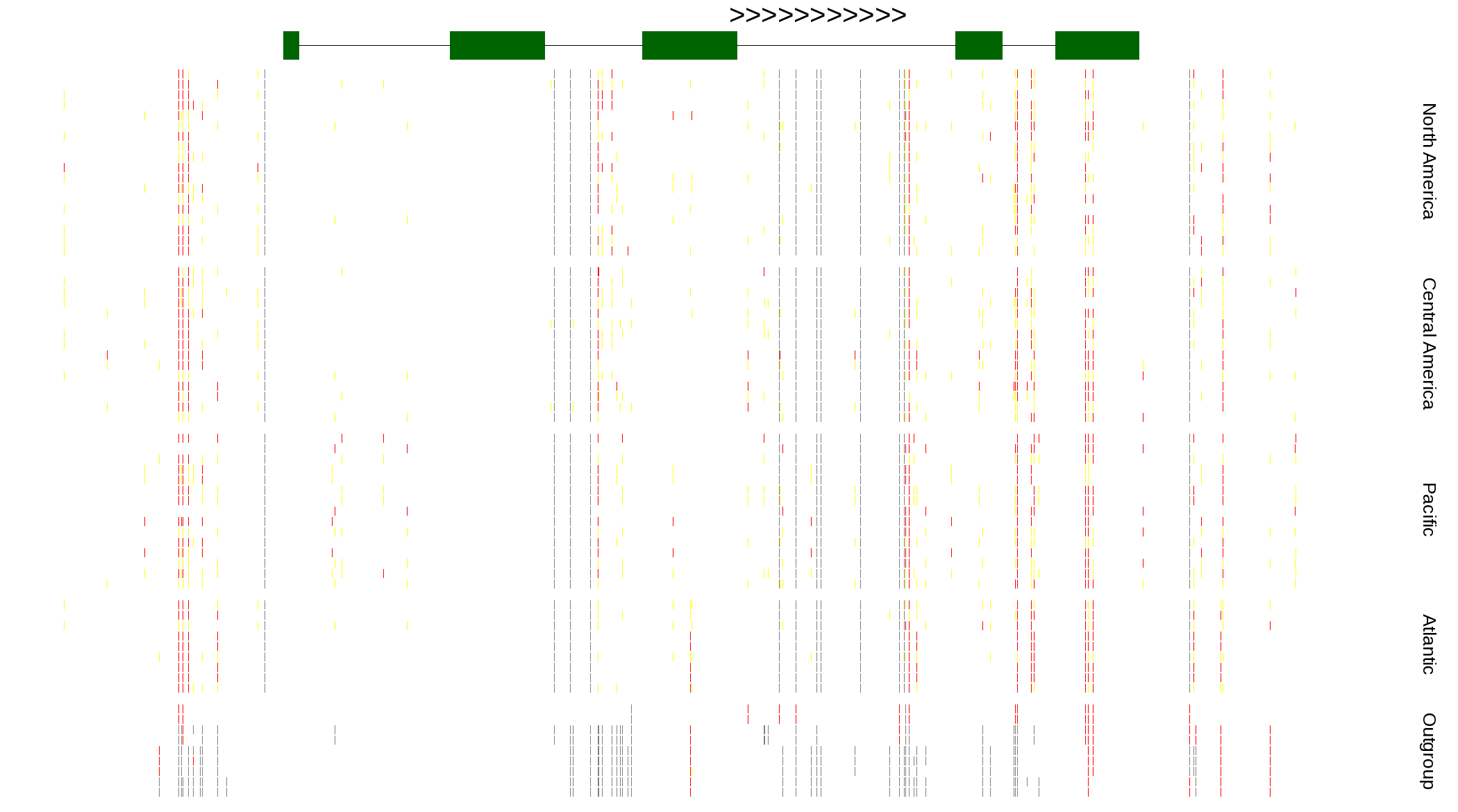
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS210094-TA
ATGGCTGGAAATACTGCTGAAGAAGTTGAGGGATCAGCACCTGCTGAGAACAAAACTACAGGGACTGTGGAGGTTCCCCCAGAGTTCTTGATAAAGCACCCGCTACAGAACACATGGAGTCTGTGGTTTTACGACAATGACAGGAATAAATCATGGGAGGAGAATCAAATTGAATTAACTTCATTTGACACAGTTGAAGATTTCTGGAGACTGTATCACCATATCAAATTGCCATCTGAGCTACGTCAAGGCCATGATTATGCTGTCTTTAAGCAAGGAATCCGTCCCATGTGGGAGGACGACGCCAACAAGATGGGTGGACGTTGGCTAATAAGTCTTGAAAAGAAGCAACGCAACTCAGATTTAGATAGGTTTTGGTTGGATGTGGTCCTGCTTCTGATTGGTGAGAATTTTGAATATGCTGAGGAAATTTGTGGCGCTGTTGTAAATGTGAGAGCAAAACTTGATAAAATTGGTGTTTGGACAGCAGACACATCGAAACAACAAGCCAATCTTGAGATTGGTAGAAAACTAAAGGAGCAACTCGGAATTCACGGAAAGATTGGATTCCAGTTACACAGAGACACTATGGTCAAACACAGTTCGGCAACTAAGAATCTTTACACTGTTTAG
>DPOGS210094-PA
MAGNTAEEVEGSAPAENKTTGTVEVPPEFLIKHPLQNTWSLWFYDNDRNKSWEENQIELTSFDTVEDFWRLYHHIKLPSELRQGHDYAVFKQGIRPMWEDDANKMGGRWLISLEKKQRNSDLDRFWLDVVLLLIGENFEYAEEICGAVVNVRAKLDKIGVWTADTSKQQANLEIGRKLKEQLGIHGKIGFQLHRDTMVKHSSATKNLYTV-