| DPOGS210963 | ||
|---|---|---|
| Transcript | DPOGS210963-TA | 435 bp |
| Protein | DPOGS210963-PA | 144 aa |
| Genomic position | DPSCF300004 - 484897-485413 | |
| RNAseq coverage | 22x (Rank: top 79%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL025000 | 4e-26 | 78.46% | |
| Bombyx | BGIBMGA006393-TA | 7e-51 | 66.67% | |
| Drosophila | ftz-PA | 2e-27 | 67.95% | |
| EBI UniRef50 | UniRef50_A9X3W6 | 2e-28 | 61.86% | Homeobox protein fushi tarazu n=1 Tax=Endeis spinosa RepID=A9X3W6_9CHEL |
| NCBI RefSeq | XP_002406404.1 | 1e-28 | 71.95% | fushi tarazu, putative [Ixodes scapularis] |
| NCBI nr blastp | gi|383849764 | 7e-28 | 65.35% | PREDICTED: uncharacterized protein LOC100875892 [Megachile rotundata] |
| NCBI nr blastx | gi|156551123 | 5e-28 | 64.36% | PREDICTED: hypothetical protein LOC100110062 [Nasonia vitripennis] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0006355 | 8.5e-26 | regulation of transcription, DNA-dependent | |
| GO:0043565 | 8.5e-26 | sequence-specific DNA binding | ||
| GO:0003700 | 8.5e-26 | sequence-specific DNA binding transcription factor activity | ||
| GO:0003677 | 7.7e-25 | DNA binding | ||
| GO:0005515 | 1.9e-22 | protein binding | ||
| GO:0005634 | 5e-06 | nucleus | ||
| KEGG pathway | ||||
| InterPro domain | [13-75] IPR001356 | 8.5e-26 | Homeobox | |
| [7-71] IPR012287 | 7.7e-25 | Homeodomain-related | ||
| [12-86] IPR009057 | 1.9e-22 | Homeodomain-like | ||
| [35-46] IPR020479 | 5.6e-09 | Homeobox, eukaryotic | ||
| [42-51] IPR000047 | 5e-06 | Helix-turn-helix motif, lambda-like repressor | ||
| Orthology group | MCL25239 | Insect specific | ||
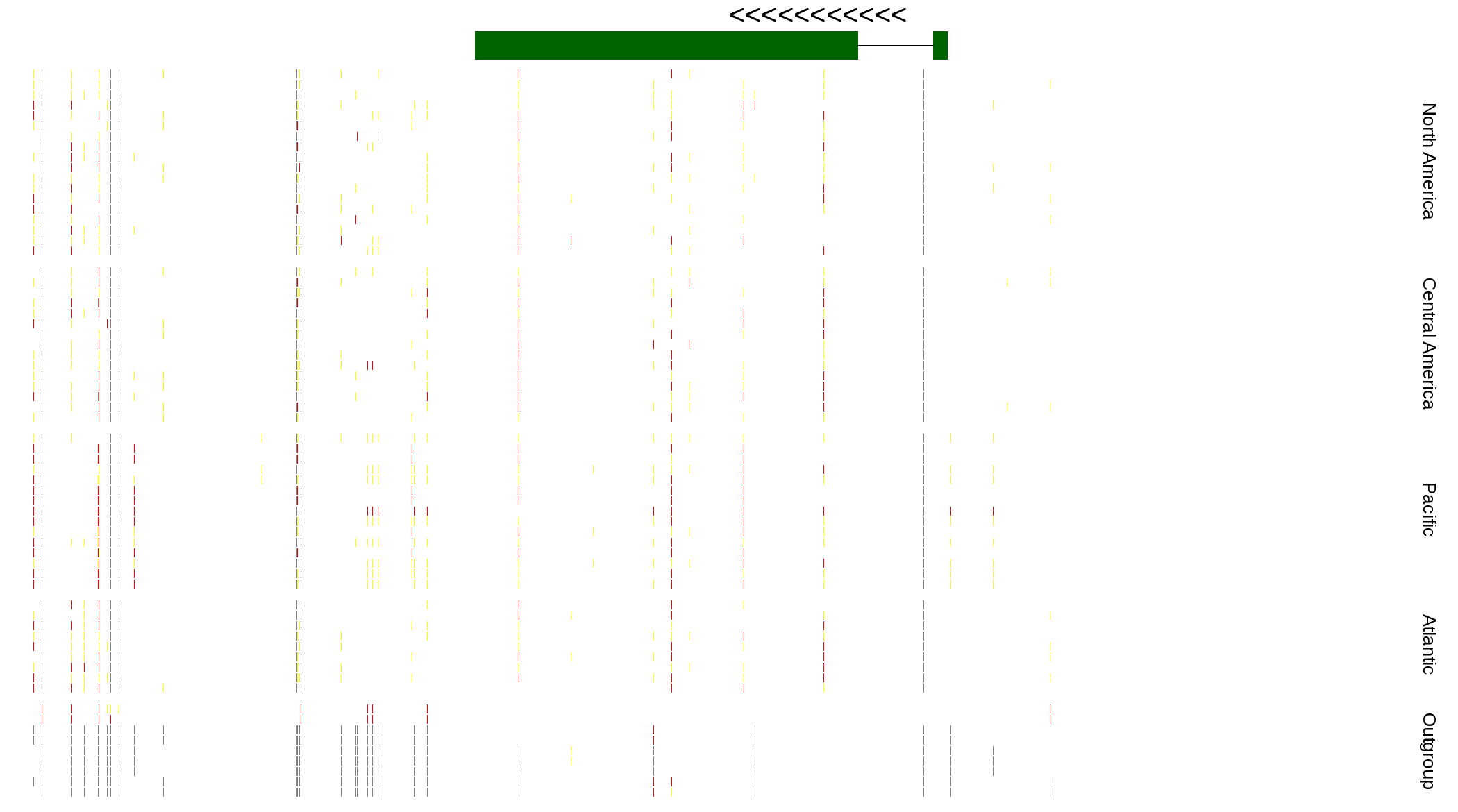
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS210963-TA
ATGAAAAGCATTGGAGGTGATGACAAAAAAGAAGGCTCGAAGCGGACCCGACAAACTTACACGAGGTTTCAAACATTGGAGCTGGAGAAAGAATTTCACTTCAACAAATATTTATCAAGACGGCGCCGGATAGAGGTTTCACACGCACTGGGTCTGACGGAGAGACAAATAAAGATATGGTTCCAGAACAGGAGAATGAAGGCAAAGAAAGATGGGAAGTTGACGAATTCACCAGATCCATTTGAAGATCTGAACAGTAAAGCAGCCAACGTTAATGACTTCGCTGATCCAAGGCAGCAAATACCAAAATTCATGGATTTCCCGAATCCCAACTATCACCTGACCGGCAATCCCATCGCAAACATGAGTCATTTAACAGGAAACCTAAATCAAACCTGCAACATACCTTACGGCGGTGTCATACCAAAAATGTGA
>DPOGS210963-PA
MKSIGGDDKKEGSKRTRQTYTRFQTLELEKEFHFNKYLSRRRRIEVSHALGLTERQIKIWFQNRRMKAKKDGKLTNSPDPFEDLNSKAANVNDFADPRQQIPKFMDFPNPNYHLTGNPIANMSHLTGNLNQTCNIPYGGVIPKM-