| DPOGS211448 | ||
|---|---|---|
| Transcript | DPOGS211448-TA | 579 bp |
| Protein | DPOGS211448-PA | 192 aa |
| Genomic position | DPSCF300223 + 43012-44275 | |
| RNAseq coverage | 1368x (Rank: top 9%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL007452 | 2e-52 | 57.14% | |
| Bombyx | BGIBMGA002156-TA | 3e-32 | 60.16% | |
| Drosophila | srp-PA | 3e-20 | 68.85% | |
| EBI UniRef50 | UniRef50_P52167-3 | 1e-25 | 49.67% | Isoform 3 of Transcription factor BCFI n=1 Tax=Bombyx mori RepID=P52167-3 |
| NCBI RefSeq | XP_001607832.1 | 3e-24 | 41.56% | PREDICTED: similar to transcription factor GATA-4 (GATA binding factor-4) [Nasonia vitripennis] |
| NCBI nr blastp | gi|603162 | 6e-26 | 50.33% | BmGATA beta isoform 3, partial [Bombyx mori] |
| NCBI nr blastx | gi|339236855 | 2e-28 | 40.93% | zinc finger protein [Trichinella spiralis] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0006355 | 1e-23 | regulation of transcription, DNA-dependent | |
| GO:0008270 | 1e-23 | zinc ion binding | ||
| GO:0003700 | 1e-23 | sequence-specific DNA binding transcription factor activity | ||
| GO:0043565 | 2.3e-20 | sequence-specific DNA binding | ||
| KEGG pathway | ||||
| InterPro domain | [33-89] IPR013088 | 1e-23 | Zinc finger, NHR/GATA-type | |
| [31-81] IPR000679 | 2.3e-20 | Zinc finger, GATA-type | ||
| Orthology group | MCL19015 | Insect specific | ||
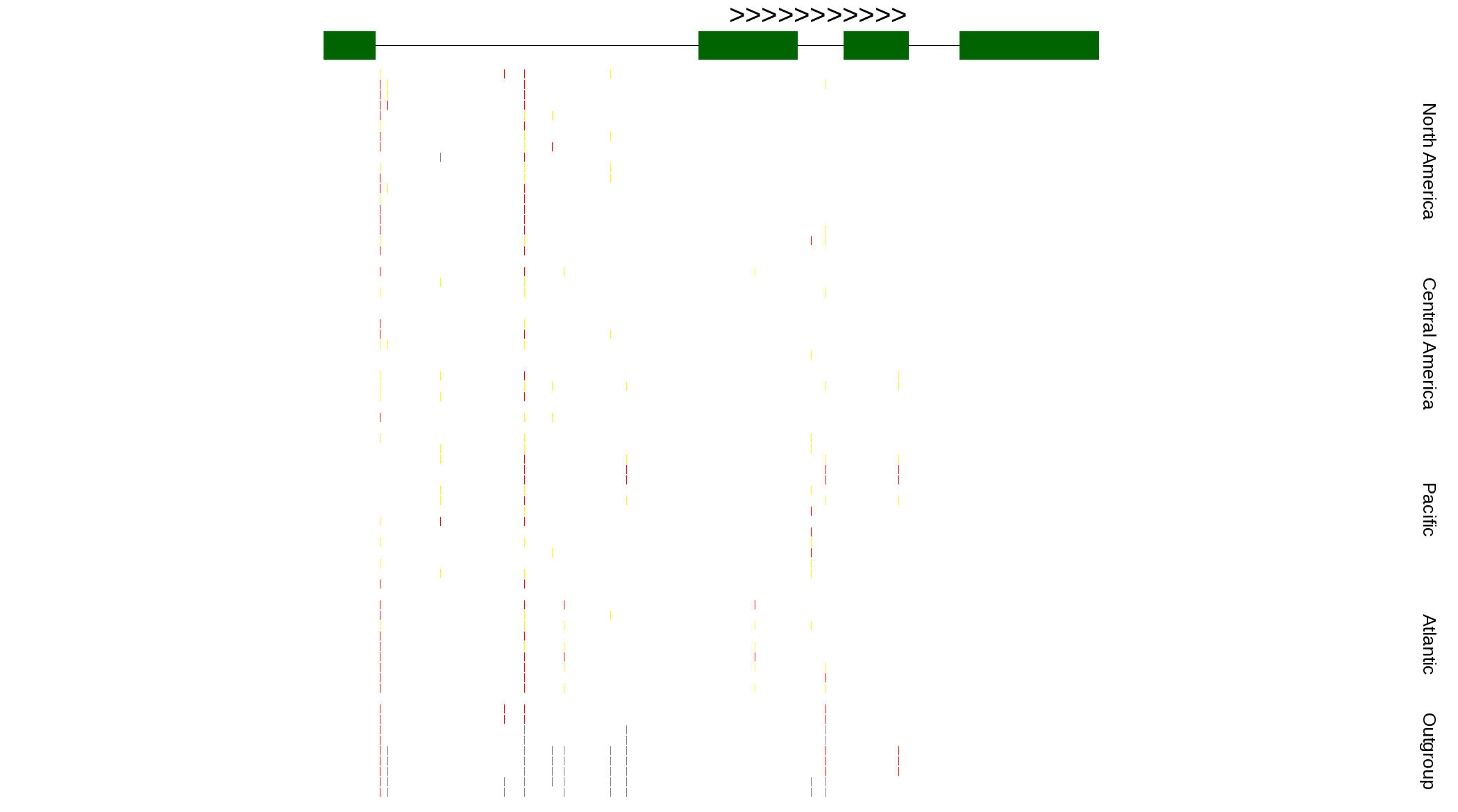
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS211448-TA
ATGCACGCCTTGCCCGCGCCCTCGCCCATCTGTGCGCCCGCGCTCGCCGTCTCCACTCCCCCTGACTACAAACAGAAGAAAGGGATGGGTACGAAGCGGCAGGGAGTGTGTTCTAACTGTGAGACCACCATCACTACTTTATGGCGTCGGAATCCGCTCGGGGAGAACGTGTGCAACGCTTGCGGGTTGTACTTCAAACTGCACGGCATCAACCGCCCGAAGAACATGAAGAAGGATTCGATCCAGACGAGAAAGCGAAAATCTAAGAACAATACGAAAACGGAGCGCAATATAAGTAAAACCACCGTTCGCTCGACTCTCAACGTAGGGACGACGGAGTTAGAGAACATATTGGATATAGGCGCCTCGTCGAGCGGTGCTCGCGGCCGTTCGCTGGGGTACTACGTGCAGCCGCACACTGTGAAACTGGAGGAGCCCGCGCCAGCCCACGCGCACCAGCAGCAGCAACAACACCAGCAACACCAACAGCAGCAGCAGGCCTACTACGACGACGAGTACCGCCGCGTCGAGCCCCAGGAGCGCCTGGAGCGGCCGACTGTGGTGTCGCTCGGCAGCTGA
>DPOGS211448-PA
MHALPAPSPICAPALAVSTPPDYKQKKGMGTKRQGVCSNCETTITTLWRRNPLGENVCNACGLYFKLHGINRPKNMKKDSIQTRKRKSKNNTKTERNISKTTVRSTLNVGTTELENILDIGASSSGARGRSLGYYVQPHTVKLEEPAPAHAHQQQQQHQQHQQQQQAYYDDEYRRVEPQERLERPTVVSLGS-