| DPOGS212277 | ||
|---|---|---|
| Transcript | DPOGS212277-TA | 438 bp |
| Protein | DPOGS212277-PA | 145 aa |
| Genomic position | DPSCF300077 + 45443-47122 | |
| RNAseq coverage | 1587x (Rank: top 8%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL002896 | 7e-40 | 65.42% | |
| Bombyx | BGIBMGA011438-TA | 1e-54 | 50.75% | |
| Drosophila | PHGPx-PD | 3e-28 | 54.00% | |
| EBI UniRef50 | UniRef50_D6WGA1 | 9e-44 | 72.22% | Glutathione peroxidase n=1 Tax=Tribolium castaneum RepID=D6WGA1_TRICA |
| NCBI RefSeq | NP_001036999.1 | 3e-53 | 50.75% | glutathione peroxidase [Bombyx mori] |
| NCBI nr blastp | gi|112983348 | 7e-52 | 50.75% | glutathione peroxidase [Bombyx mori] |
| NCBI nr blastx | gi|112983348 | 1e-51 | 51.26% | glutathione peroxidase [Bombyx mori] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0006979 | 6e-41 | response to oxidative stress | |
| GO:0004602 | 6e-41 | glutathione peroxidase activity | ||
| GO:0055114 | 6e-41 | oxidation-reduction process | ||
| KEGG pathway | tca:658456 | 1e-43 | ||
| K00432 (E1.11.1.9) | maps-> | Arachidonic acid metabolism | ||
| Glutathione metabolism | ||||
| InterPro domain | [45-139] IPR000889 | 6e-41 | Glutathione peroxidase | |
| [38-143] IPR012336 | 8.1e-38 | Thioredoxin-like fold | ||
| Orthology group | ||||
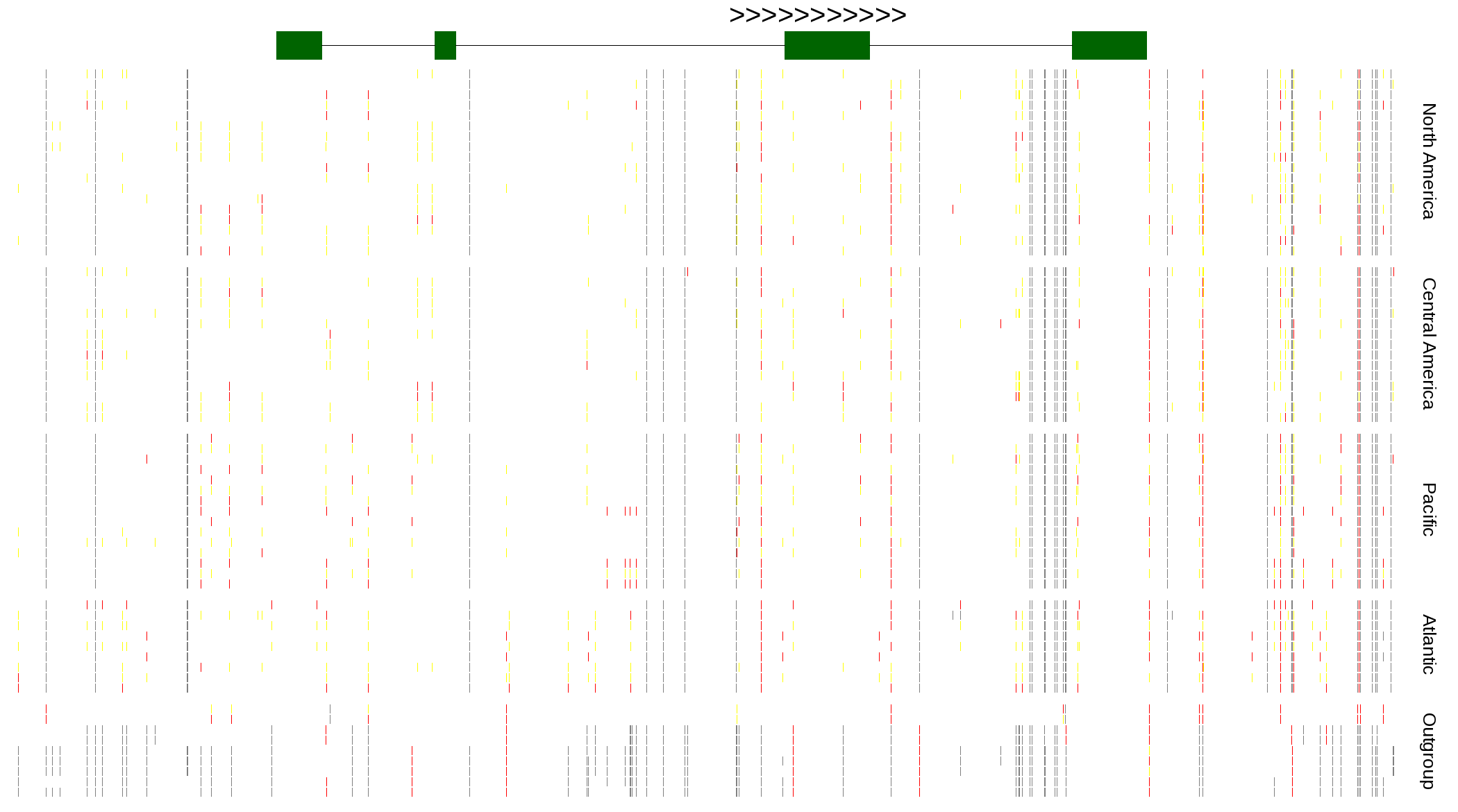
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS212277-TA
ATGACAATCGCGTATAGAACCTTATCGAAGCTTGTTGTACCTGCTCTCGGTAATTTATTTTGTGTAGCACGAGTAAATCTAAGTACCATAAAAATGGCTAGCAATAACCCAGATTACAAAAGTGCTACAAGTCTTCGTATTTTGGCGTTTCCCTGCAACCAATTTGCTGGCCAAGAGCCTGGGAACTCTGAAGACATTGTCTGTTTCATGAACGAACGCAAAGTAAACTTTGACATGTTCGAAAAGATAGATGTGAATGGCGATTCTGCGCACCCATTATGGAAATTTCTGAAGCACAAGCAGGGTGGAACTTTGGGCAATTTTATCAAGTGGAATTTCACTAAATTCATTGTCGACAAAAATGGTGTACCGGTCGAAAGACATGGCCCGAACACCGACCCTATAAACTTGGTGAAGTCTCTTGAGAATTACTGGTAA
>DPOGS212277-PA
MTIAYRTLSKLVVPALGNLFCVARVNLSTIKMASNNPDYKSATSLRILAFPCNQFAGQEPGNSEDIVCFMNERKVNFDMFEKIDVNGDSAHPLWKFLKHKQGGTLGNFIKWNFTKFIVDKNGVPVERHGPNTDPINLVKSLENYW-