| DPOGS212628 | ||
|---|---|---|
| Transcript | DPOGS212628-TA | 720 bp |
| Protein | DPOGS212628-PA | 239 aa |
| Genomic position | DPSCF300245 + 300946-302497 | |
| RNAseq coverage | 0x (Rank: top 98%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL002469 | 3e-41 | 38.05% | |
| Bombyx | BGIBMGA005196-TA | 3e-36 | 39.56% | |
| Drosophila | CG17292-PB | 2e-25 | 33.98% | |
| EBI UniRef50 | UniRef50_Q173Z0 | 1e-29 | 34.43% | Vitellogenin, putative n=5 Tax=Culicidae RepID=Q173Z0_AEDAE |
| NCBI RefSeq | XP_553025.3 | 2e-32 | 36.32% | AGAP009104-PA [Anopheles gambiae str. PEST] |
| NCBI nr blastp | gi|158299835 | 3e-31 | 36.32% | AGAP009104-PA [Anopheles gambiae str. PEST] |
| NCBI nr blastx | gi|158299835 | 3e-30 | 36.32% | AGAP009104-PA [Anopheles gambiae str. PEST] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0003824 | 3.2e-33 | catalytic activity | |
| GO:0006629 | 3.2e-33 | lipid metabolic process | ||
| KEGG pathway | dre:445105 | 6e-16 | ||
| K01066 (E3.1.1.-) | maps-> | 2,4-Dichlorobenzoate degradation | ||
| Tropane, piperidine and pyridine alkaloid biosynthesis | ||||
| InterPro domain | [30-235] IPR000734 | 3.2e-33 | Lipase | |
| [54-232] IPR013818 | 4.3e-28 | Lipase, N-terminal | ||
| Orthology group | MCL27837 | Specific divergent | ||
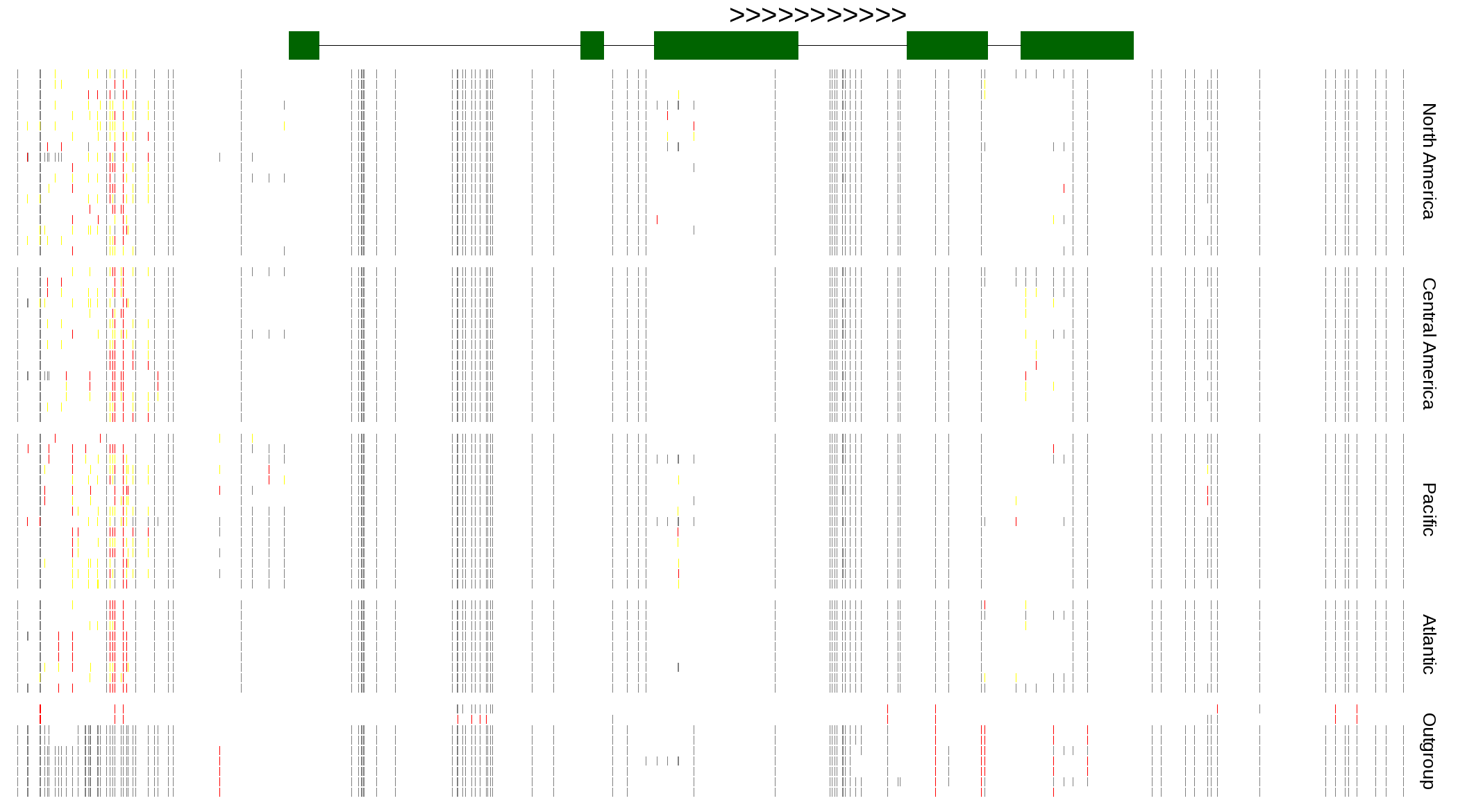
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS212628-TA
ATGACTGTTGAAAGACAAAGAAAATGTCTGCTTTTGATGATATTTTTTGCATTCGGCATTTGCTGTGCGGATGCTGCATATCTGAGAATTTATTGCGGAGAGAACTTTGACGAATACGATGAGTTTGATTTGGAATCTCCGCTGGGACTTTTGGATTCGACTGGATACGATGAAAGTCGACATACTGTTATCTACACATTCGGTTTCCAAGGAAAAGCTAATGGACAGTCTGTAAAAACGATCGTAGAAACTTATTTGAGAATCGGCAATATCAATATCATTCTCTTTAATTGGGAAAAGGAAGCAACCACACCTCTAGGCGCAATTTCATATGGAACCATAGTTGCGAAAACTGTCAATAAGCTTGGCACAAAACTGGGAGACGTCTTGGTCTCATTAGTATCAGCAGGCCTGGACATTAATAAGGTTCATTTAATTGGCTTTTCTCTTGGAGCACAACTTTTTGGATACACAGGGCGGCAAGTGATAAGGAATAATTTCCAAATACAGAGAATTACAGGTCTAGATCCAGCTGGTCCTTTGTATGATGGAAAGTTCTTCGAATCTCTAGACGAAGACAGCGCTAGTTTTGTCGACGTTTTTCATACTAACCCAAGCGGGCTAGGTAGTCATAAGTCACAAGCTACAATTGACATCTGGTTTAATTGTGAACGGAAATATCAACCAGGGTGTGAAGTTGAAGATGACCCAGGTACTTGA
>DPOGS212628-PA
MTVERQRKCLLLMIFFAFGICCADAAYLRIYCGENFDEYDEFDLESPLGLLDSTGYDESRHTVIYTFGFQGKANGQSVKTIVETYLRIGNINIILFNWEKEATTPLGAISYGTIVAKTVNKLGTKLGDVLVSLVSAGLDINKVHLIGFSLGAQLFGYTGRQVIRNNFQIQRITGLDPAGPLYDGKFFESLDEDSASFVDVFHTNPSGLGSHKSQATIDIWFNCERKYQPGCEVEDDPGT-