| DPOGS212651 | ||
|---|---|---|
| Transcript | DPOGS212651-TA | 1341 bp |
| Protein | DPOGS212651-PA | 446 aa |
| Genomic position | DPSCF300319 + 60900-65807 | |
| RNAseq coverage | 16x (Rank: top 81%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL013626 | 2e-120 | 51.44% | |
| Bombyx | BGIBMGA007391-TA | 2e-119 | 45.05% | |
| Drosophila | CG6321-PA | 3e-27 | 26.97% | |
| EBI UniRef50 | UniRef50_E0V9A9 | 3e-76 | 41.99% | Kynurenine/alpha-aminoadipate aminotransferase, putative n=3 Tax=Coelomata RepID=E0V9A9_PEDHC |
| NCBI RefSeq | XP_973433.1 | 2e-88 | 40.89% | PREDICTED: similar to Kynurenine/alpha-aminoadipate aminotransferase mitochondrial precursor (KAT/AadAT) (Kynurenine--oxoglutarate transaminase II) (Kynurenine aminotransferase II) (Kynurenine--oxoglutarate aminotransferase II) (2-aminoadipate transaminase) ( [Tribolium castaneum] |
| NCBI nr blastp | gi|91086977 | 4e-87 | 40.89% | PREDICTED: similar to Kynurenine/alpha-aminoadipate aminotransferase mitochondrial precursor (KAT/AadAT) (Kynurenine--oxoglutarate transaminase II) (Kynurenine aminotransferase II) (Kynurenine--oxoglutarate aminotransferase II) (2-aminoadipate transaminase) ( [Tribolium castaneum] |
| NCBI nr blastx | gi|91086977 | 4e-85 | 41.13% | PREDICTED: similar to Kynurenine/alpha-aminoadipate aminotransferase mitochondrial precursor (KAT/AadAT) (Kynurenine--oxoglutarate transaminase II) (Kynurenine aminotransferase II) (Kynurenine--oxoglutarate aminotransferase II) (2-aminoadipate transaminase) ( [Tribolium castaneum] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0003824 | 3.7e-47 | catalytic activity | |
| GO:0030170 | 3.7e-47 | pyridoxal phosphate binding | ||
| GO:0016769 | 1.5e-24 | transferase activity, transferring nitrogenous groups | ||
| GO:0009058 | 1.5e-24 | biosynthetic process | ||
| KEGG pathway | tca:662228 | 6e-88 | ||
| K00816 (E2.6.1.7) | maps-> | Tryptophan metabolism | ||
| K00825 (E2.6.1.39) | maps-> | Lysine degradation | ||
| Lysine biosynthesis | ||||
| InterPro domain | [42-435] IPR015424 | 1.6e-71 | Pyridoxal phosphate-dependent transferase, major domain | |
| [99-317] IPR015421 | 3.7e-47 | Pyridoxal phosphate-dependent transferase, major region, subdomain 1 | ||
| [98-428] IPR004839 | 1.5e-24 | Aminotransferase, class I/classII | ||
| [318-434] IPR015422 | 2.6e-07 | Pyridoxal phosphate-dependent transferase, major region, subdomain 2 | ||
| Orthology group | MCL23357 | Specific divergent | ||
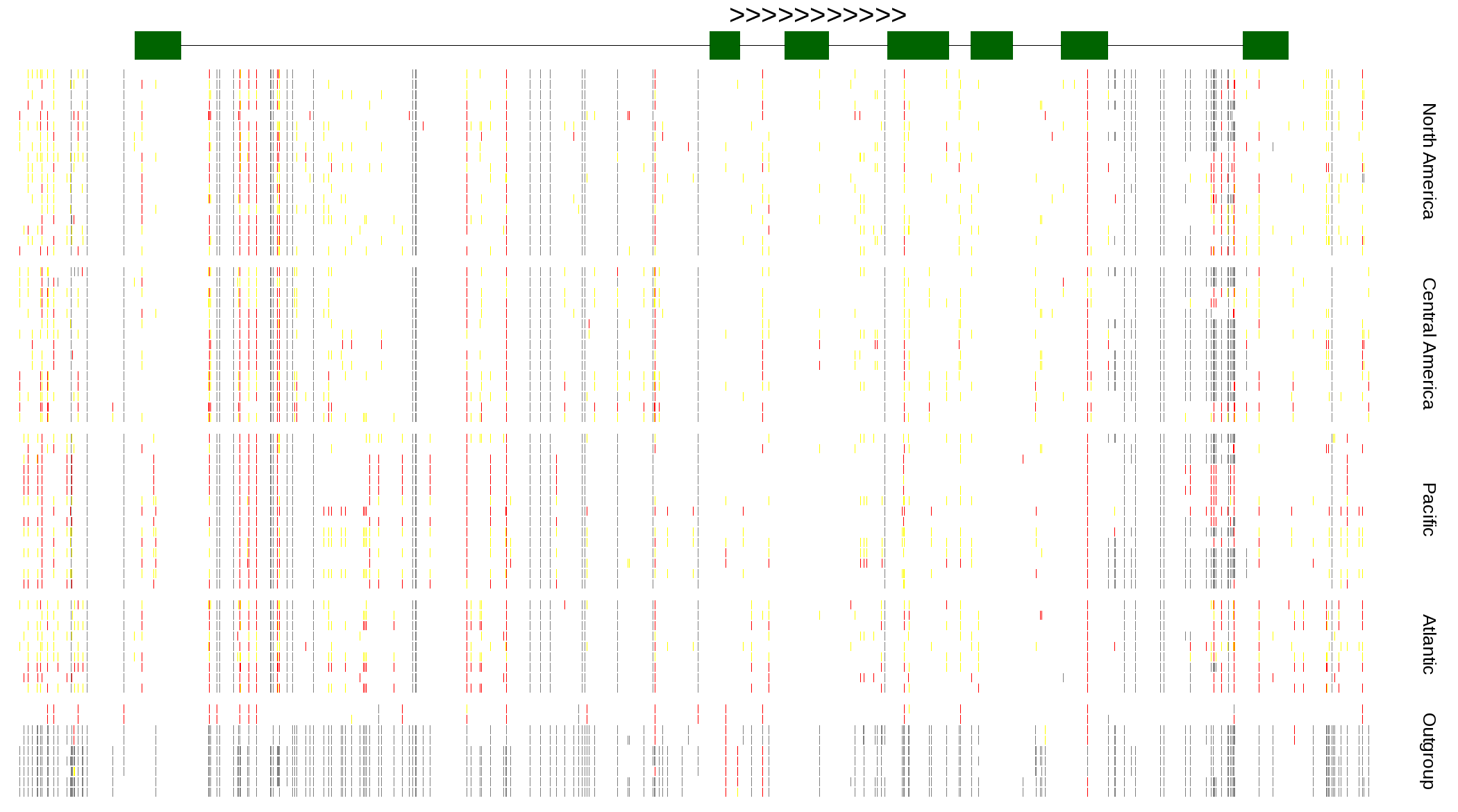
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS212651-TA
ATGTTGAATCTATTCGAAAAACGTACAATAATGTGTGGCCTGCGAAGATATTCTGAAAAAGCGAAAAATTTGTACAAGTTCTTCTGTGAGGAAGAAAACGAGCACCAGAAATACAGAACTCTCGATGAGAAGGACTATGTGAAATTTTTGAGTAAAAGAGCGCTAACGTTTAAACCTTCGGATACGAGACAATTGAGAGAAGGTATGCCCAATGAAAAAACGTTTCCGTTTACTAACCTCGAACTCACCACAAGAAGTGGTGACAAAATATCAATAGAAGGACACCAGTTGGACGAGGCACTGCAGTATATACCGCCGAGAGGATTGCCGTGTCTACATCACGAGCTAAATAAATTTCAACAAGAGCTGCATCGTCCGCCTCCTCTCCCGCGTAGTCTATTACTCACCAACGGAGCCCAGCACGGCATTCATCTTTGTACTGAACTGCTTGTGGATCACGGTGATCCCGTTATGTCCACTGAATACTCGTACATTGGCTTGCATTGTATTTTAAAACCTTATAATGTGGAATATATCGGAATCAAAGAAGATAAAGACGGTTTGATTCCAGAAGAATTAGAATCAGTGTTAAATACAAGAATGAAACAAGGACTCAAAATGCCAAGAGTATTGTTTGTTGTTCCGACAGTCAGCAACCCTGCTGGTTTCCTGTTATCTGAGAATAGGAAAAGAAAAATTTACGAGATAGCTTGCAAATACAATTTAATAATTGTAGAGGATGAAGTTTATATGTTCTTGAATTACACTGATACGGTAGTACCATCTTTCCTGTCGCTGGATACAACAGGTCGTGTGATGCGCATAGATTCCATCTCGAAGGTTGTTAGCGCTGGGTTGAGAGTGGGCTGGTTGACGGGACCTGAACCTCTCATCAGATATACTGAAATGGCTTCATTTTCGCAAATGCTACATCCTTGCTCTTTATCTCAGTCAATAGCGTACAGTCTTATGACTGACCGAAATCGCTTAACAACTCACCTACTAAGCACTATAAAGTTTTACAAAAAACAAAAAGATTGTGTAAATAATGCTTTGAAGAAGATAGAAGGTCTGATAGAATGGGAGGATCCGGCCGGGGGCTACTTCCACTGGGTAAAGATACGTGGACTTGAGGACGTGCGTAACATGGTTTTCAACACCGCCTTCAAACATGGCTTGATGTTAGAACCAGGTCACAATTTCAGTTACGACACTGAGAAGCCATCTCCGTACATACGACTTACATTCACAAGAATGAACTTGGAACAACTAGACGATAACGTCAACACGATTCGTGATATTATCATAGAAGAAAAAAAAATACAACAGAAAAAATGTTAG
>DPOGS212651-PA
MLNLFEKRTIMCGLRRYSEKAKNLYKFFCEEENEHQKYRTLDEKDYVKFLSKRALTFKPSDTRQLREGMPNEKTFPFTNLELTTRSGDKISIEGHQLDEALQYIPPRGLPCLHHELNKFQQELHRPPPLPRSLLLTNGAQHGIHLCTELLVDHGDPVMSTEYSYIGLHCILKPYNVEYIGIKEDKDGLIPEELESVLNTRMKQGLKMPRVLFVVPTVSNPAGFLLSENRKRKIYEIACKYNLIIVEDEVYMFLNYTDTVVPSFLSLDTTGRVMRIDSISKVVSAGLRVGWLTGPEPLIRYTEMASFSQMLHPCSLSQSIAYSLMTDRNRLTTHLLSTIKFYKKQKDCVNNALKKIEGLIEWEDPAGGYFHWVKIRGLEDVRNMVFNTAFKHGLMLEPGHNFSYDTEKPSPYIRLTFTRMNLEQLDDNVNTIRDIIIEEKKIQQKKC-