| DPOGS213100 | ||
|---|---|---|
| Transcript | DPOGS213100-TA | 630 bp |
| Protein | DPOGS213100-PA | 209 aa |
| Genomic position | DPSCF300016 + 52590-58869 | |
| RNAseq coverage | 543x (Rank: top 23%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL006558 | 2e-84 | 65.22% | |
| Bombyx | BGIBMGA007860-TA | 4e-50 | 61.59% | |
| Drosophila | CG9362-PA | 2e-49 | 42.65% | |
| EBI UniRef50 | UniRef50_Q1HPS2 | 6e-77 | 62.68% | Glutathione transferase zeta n=4 Tax=Obtectomera RepID=Q1HPS2_BOMMO |
| NCBI RefSeq | NP_001040453.1 | 1e-77 | 62.68% | glutathione S-transferae zeta 2 [Bombyx mori] |
| NCBI nr blastp | gi|224471387 | 1e-76 | 62.68% | zeta-class glutathione S-transferase [Bombyx mori] |
| NCBI nr blastx | gi|224471387 | 4e-73 | 62.68% | zeta-class glutathione S-transferase [Bombyx mori] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0009072 | 9e-62 | aromatic amino acid family metabolic process | |
| GO:0003824 | 9e-62 | catalytic activity | ||
| GO:0005737 | 9e-62 | cytoplasm | ||
| GO:0005515 | 2.7e-13 | protein binding | ||
| KEGG pathway | dre:436754 | 4e-51 | ||
| K01800 (E5.2.1.2, maiA) | maps-> | Styrene degradation | ||
| Tyrosine metabolism | ||||
| InterPro domain | [7-207] IPR005955 | 9e-62 | Maleylacetoacetate isomerase | |
| [100-207] IPR010987 | 1.1e-29 | Glutathione S-transferase, C-terminal-like | ||
| [2-99] IPR012336 | 2.8e-25 | Thioredoxin-like fold | ||
| [8-79] IPR004045 | 2.7e-13 | Glutathione S-transferase, N-terminal | ||
| Orthology group | MCL25398 | Lepidoptera specific | ||
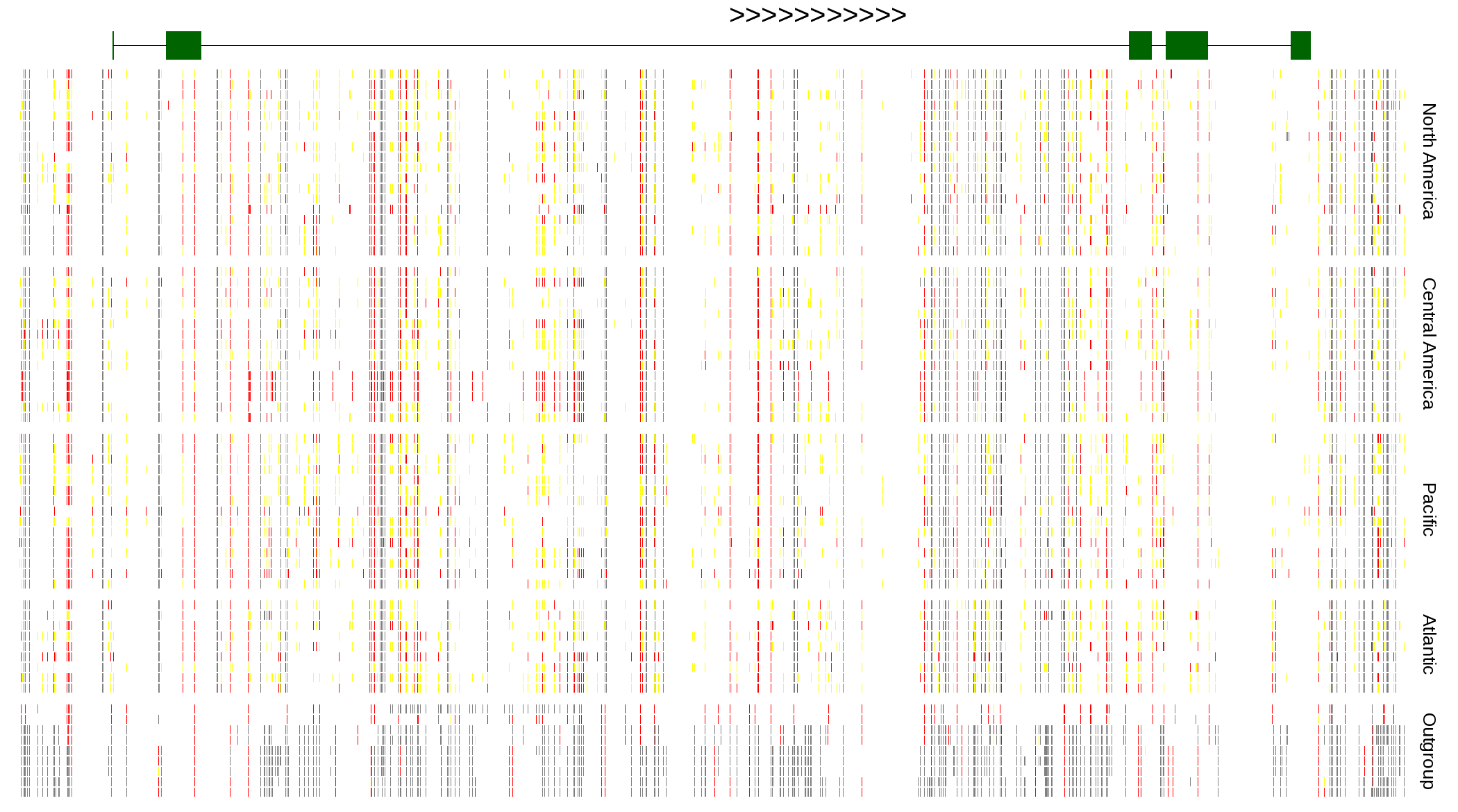
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS213100-TA
ATGGCGGAAACACGTGCTATACTGTACGGGTTTTGGGCGTCATCCTGTACGTGGCGCGTCAGAGCAGCCTTACACTTCAAAGGCATTGCGTTTGAGGAGAAATCAGTCGATATTGTTACTGAGCAAAAACAACTCACAGATGAATATCGATATATTAACCCATCCCAAAAAGTTCCAGCACTAGTAATGAATGGTGAAACAATTGTGGAATCGATGGCAATTATACAATATGTGGAAGAAATAAAACCAGAAGCAAGCCTTGTTCCGACAACACCAATTCTGAGAGCTCGTATGAGAGAAATTTGTGAGACTATTGTAAGCGGAATCCAGCCCCTGCAAAACATCGGTCTAAAGAGGCGTTTCGATTCAGAGCATAAGTTCAAAGAATTTGCCGAATACTTTACGTCGCGTGGTCTAGAAAGTGTTGAGGAACTGCTTAAAAAAACCGCCGGCCGCTTTTGTATTGGCGATCAAATAACCGTTGCCGACTTGTGTCTTGTACCACAAGTTTATAACGGGATTGTGAGATATAAAATGAACATGGAAAAGTTCCCAATTGTTTCCAGTGTATACGAGCATCTCTTGAAAGAAAAAACTTTTATCGAGACTCATCCAAAGAACATCAAATGA
>DPOGS213100-PA
MAETRAILYGFWASSCTWRVRAALHFKGIAFEEKSVDIVTEQKQLTDEYRYINPSQKVPALVMNGETIVESMAIIQYVEEIKPEASLVPTTPILRARMREICETIVSGIQPLQNIGLKRRFDSEHKFKEFAEYFTSRGLESVEELLKKTAGRFCIGDQITVADLCLVPQVYNGIVRYKMNMEKFPIVSSVYEHLLKEKTFIETHPKNIK-