| DPOGS213142 | ||
|---|---|---|
| Transcript | DPOGS213142-TA | 684 bp |
| Protein | DPOGS213142-PA | 227 aa |
| Genomic position | DPSCF300016 + 1002943-1011825 | |
| RNAseq coverage | 1925x (Rank: top 6%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL010347 | 8e-109 | 95.85% | |
| Bombyx | BGIBMGA007901-TA | 3e-109 | 87.38% | |
| Drosophila | Peritrophin-A-PA | 9e-88 | 72.22% | |
| EBI UniRef50 | UniRef50_G6D270 | 5e-132 | 100.00% | Cuticular protein analogous to peritrophins 3-D1 n=12 Tax=Endopterygota RepID=G6D270_DANPL |
| NCBI RefSeq | NP_001161908.1 | 2e-96 | 74.89% | cuticular protein analogous to peritrophins 3-D1 [Tribolium castaneum] |
| NCBI nr blastp | gi|332375180 | 9e-97 | 74.45% | unknown [Dendroctonus ponderosae] |
| NCBI nr blastx | gi|332375180 | 3e-105 | 75.45% | unknown [Dendroctonus ponderosae] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0008061 | 8.3e-13 | chitin binding | |
| GO:0006030 | 8.3e-13 | chitin metabolic process | ||
| GO:0005576 | 8.3e-13 | extracellular region | ||
| KEGG pathway | tca:662504 | 9e-06 | ||
| K01873 (VARS, valS) | maps-> | Aminoacyl-tRNA biosynthesis | ||
| Valine, leucine and isoleucine biosynthesis | ||||
| InterPro domain | [20-86] IPR002557 | 8.3e-13 | Chitin binding domain | |
| Orthology group | MCL14697 | Insect specific | ||
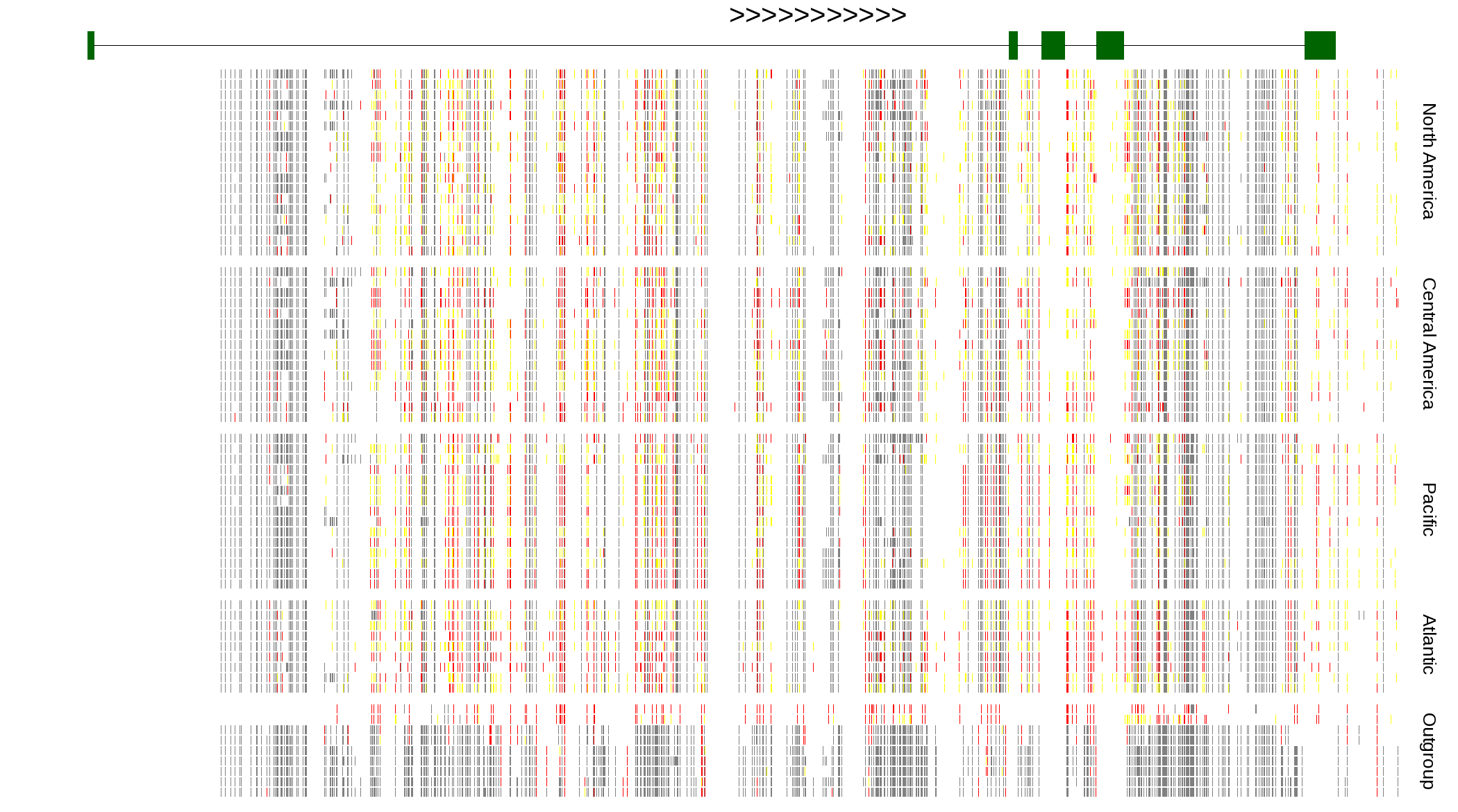
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS213142-TA
ATGATAAGTTCCGTATTTGCTGTTTTAACACTTTGCGTTGTATACGCGCACGCAGGGATCCTTCTTCCACATGCTCCAACCTGTCCGGAGCACTATGGAGTTCAGGCATATGCCCACCCCGAGTTATGTGATCAATTCTTTTTGTGCACAAACGGCACACTAACCGTCGAAACATGCGAGAACGGCCTTCTGTTTGACGGCAAGGGCGCAGTACACAATCATTGCAACTACCACTGGGCCGTCGACTGCGGTGAAAGGAAAGCTGACTTGACACCATACTCTACTCCTGGTTGCGAATACCAGTTCGGTATATACCCAGATAGCGCTGAATGTTCAACAAGCTACATTAAATGCGCTTTCGGCATCCCCCATCAAGAACCTTGCACCCCTGGTCTAGTATACGACGAGCGTATCCATGGTTGCAACTGGCCTGATCTCCTGCAACCTTTCTGTAACCCTGAAGCTGTCGTTGGCTTCAAGTGTCCAACTAAAGTTCCCGCCAATACCCAGTCAGCCAAGTTCTGGCCTTTCCCTCGTTTCCCGGTGCCCGGAGACTGCCACAGACTGATCACATGCGTAGAGGGAAACCCTCGACTGATCACCTGCGGAGAAGGAAAAGTTTTCGACGACCAGAACCTCACTTGTGAAGATCCCGAATTAGTGCCACACTGTGCACACGCTTAA
>DPOGS213142-PA
MISSVFAVLTLCVVYAHAGILLPHAPTCPEHYGVQAYAHPELCDQFFLCTNGTLTVETCENGLLFDGKGAVHNHCNYHWAVDCGERKADLTPYSTPGCEYQFGIYPDSAECSTSYIKCAFGIPHQEPCTPGLVYDERIHGCNWPDLLQPFCNPEAVVGFKCPTKVPANTQSAKFWPFPRFPVPGDCHRLITCVEGNPRLITCGEGKVFDDQNLTCEDPELVPHCAHA-