| DPOGS213165 | ||
|---|---|---|
| Transcript | DPOGS213165-TA | 627 bp |
| Protein | DPOGS213165-PA | 208 aa |
| Genomic position | DPSCF300114 - 479528-481822 | |
| RNAseq coverage | 327x (Rank: top 35%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL017080 | 3e-23 | 43.79% | |
| Bombyx | BGIBMGA007350-TA | 2e-29 | 72.09% | |
| Drosophila | Vti1-PA | 2e-09 | 40.91% | |
| EBI UniRef50 | UniRef50_E2AHH8 | 1e-13 | 45.05% | Vesicle transport through interaction with t-SNAREs-like protein 1A n=10 Tax=Coelomata RepID=E2AHH8_CAMFO |
| NCBI RefSeq | XP_623763.2 | 1e-16 | 45.45% | PREDICTED: similar to CG3279-PA [Apis mellifera] |
| NCBI nr blastp | gi|380021429 | 2e-15 | 45.45% | PREDICTED: LOW QUALITY PROTEIN: vesicle transport through interaction with t-SNAREs homolog 1A-like [Apis florea] |
| NCBI nr blastx | gi|380021429 | 2e-16 | 45.10% | PREDICTED: LOW QUALITY PROTEIN: vesicle transport through interaction with t-SNAREs homolog 1A-like [Apis florea] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0016020 | 4.3e-16 | membrane | |
| GO:0016192 | 4.3e-16 | vesicle-mediated transport | ||
| GO:0006886 | 6.5e-12 | intracellular protein transport | ||
| KEGG pathway | ame:551363 | 4e-16 | ||
| K08493 (VTI1) | maps-> | SNARE interactions in vesicular transport | ||
| InterPro domain | [6-88] IPR010989 | 4.3e-16 | t-SNARE | |
| [13-83] IPR007705 | 6.5e-12 | Vesicle transport v-SNARE, N-terminal | ||
| Orthology group | MCL34966 | Lepidoptera specific | ||
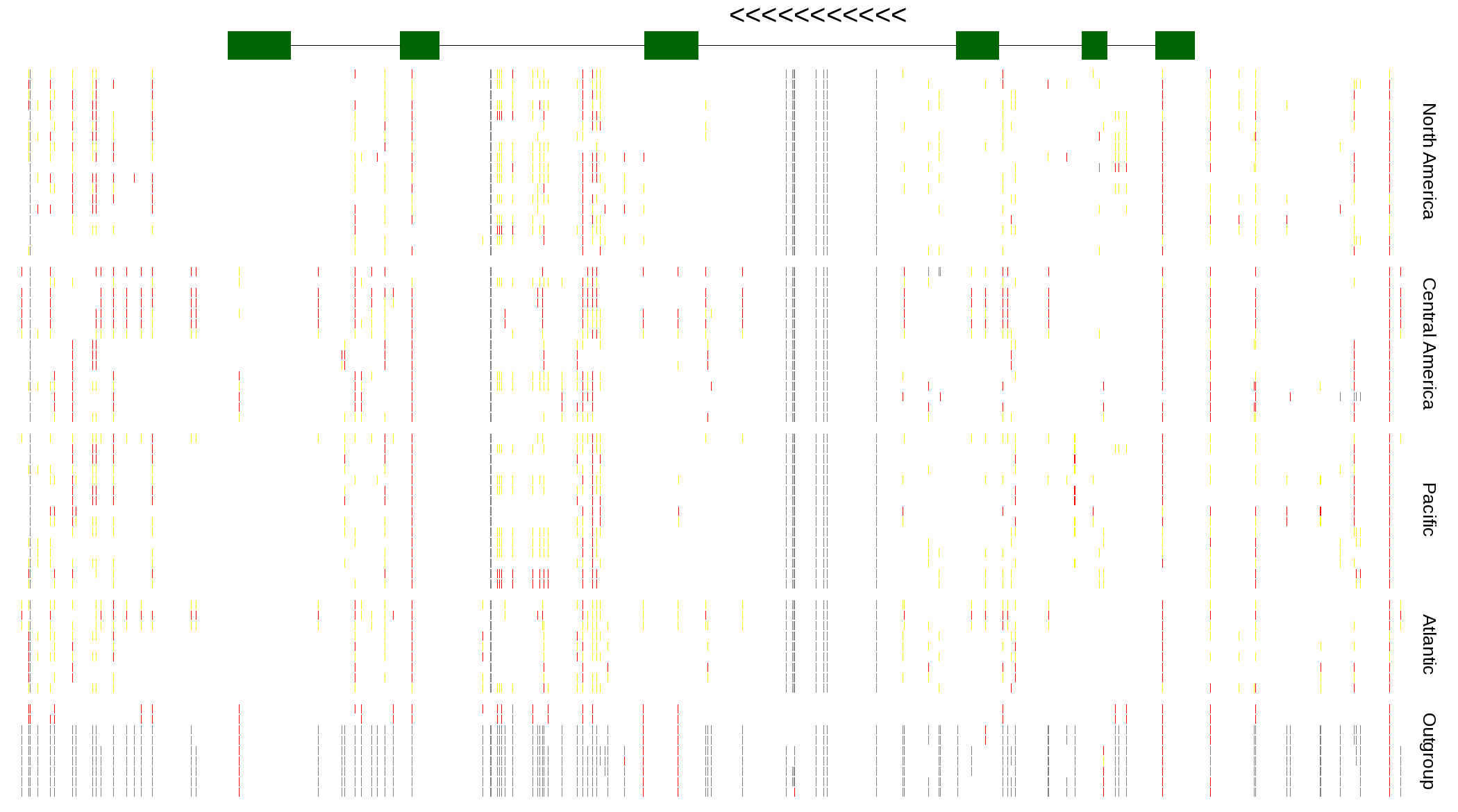
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS213165-TA
ATGTCGACATTAATTCAATCATATGAACAACAGTATTCTATTCTTACGGCTGAAATTACTGCAAAAATAGGGCGACTGAAGACAGCTCATGAAGATGACCGTGATCAATTATCAAGAGAGATTCAGTCAAATTTCGAGGAGGCCAATGATTTGTTGGAGCAATTGGAATTAGAATCTCGCGGCTCCGGTGGAGGGTCAAGAGTTCCGGCCTACCGTGCTGAACTGCAACGAGTCAGGGATGAGTTCAGATCAGCTCTACGTGAGACGGATGAACAGCTGTCTCGCTCTGGACGTCTCATGAACACTATGGTGACCCGAGCTCTGCAGCAGCGTGCTGTTCTAGCCGCTGTGCTGCTAGTCTTGGTCATTCTAGCAGCACTAGGGGCCCGACAATTGACTATGAGCGCCTGCGAAAAATATGCGAGCAATAACGCAAGGACTCGGGACGTGGTGAAGATAGGGTCTGTGCGTAACGAAGAAGTGAAGGACGAGCCATTGGTCCATTTTCTGGTATATTGGCTGTATATAGCATGCAGCAAAGTACTGCCTTCGGGCCTTCGGGTCTGGTTTGTGCTACGATGTCTGTGGGCCTTTCTCTGCCACAGGTACGAGAGCCTGAGTGGCTAA
>DPOGS213165-PA
MSTLIQSYEQQYSILTAEITAKIGRLKTAHEDDRDQLSREIQSNFEEANDLLEQLELESRGSGGGSRVPAYRAELQRVRDEFRSALRETDEQLSRSGRLMNTMVTRALQQRAVLAAVLLVLVILAALGARQLTMSACEKYASNNARTRDVVKIGSVRNEEVKDEPLVHFLVYWLYIACSKVLPSGLRVWFVLRCLWAFLCHRYESLSG-