| DPOGS213607 | ||
|---|---|---|
| Transcript | DPOGS213607-TA | 741 bp |
| Protein | DPOGS213607-PA | 246 aa |
| Genomic position | DPSCF300033 + 755806-758030 | |
| RNAseq coverage | 119x (Rank: top 58%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL013677 | 6e-122 | 86.23% | |
| Bombyx | BGIBMGA011669-TA | 1e-134 | 92.31% | |
| Drosophila | CG18177-PA | 1e-69 | 56.17% | |
| EBI UniRef50 | UniRef50_F4W499 | 8e-73 | 60.79% | N-acetyltransferase 15 n=5 Tax=Formicidae RepID=F4W499_ACREC |
| NCBI RefSeq | XP_624819.1 | 5e-73 | 62.11% | PREDICTED: similar to CG18177-PB, isoform B [Apis mellifera] |
| NCBI nr blastp | gi|307192783 | 7e-73 | 62.11% | N-acetyltransferase UNQ2771/PRO7155-like protein [Harpegnathos saltator] |
| NCBI nr blastx | gi|307192783 | 6e-74 | 62.56% | N-acetyltransferase UNQ2771/PRO7155-like protein [Harpegnathos saltator] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0008152 | 6.6e-08 | metabolic process | |
| GO:0008080 | 6.6e-08 | N-acetyltransferase activity | ||
| KEGG pathway | smm:Smp_131280 | 2e-08 | ||
| K14021 (BAK, BAK1) | maps-> | Protein processing in endoplasmic reticulum | ||
| InterPro domain | [27-204] IPR016181 | 7e-21 | Acyl-CoA N-acyltransferase | |
| [77-178] IPR000182 | 6.6e-08 | GCN5-related N-acetyltransferase (GNAT) domain | ||
| Orthology group | MCL15196 | Single-copy universal gene | ||
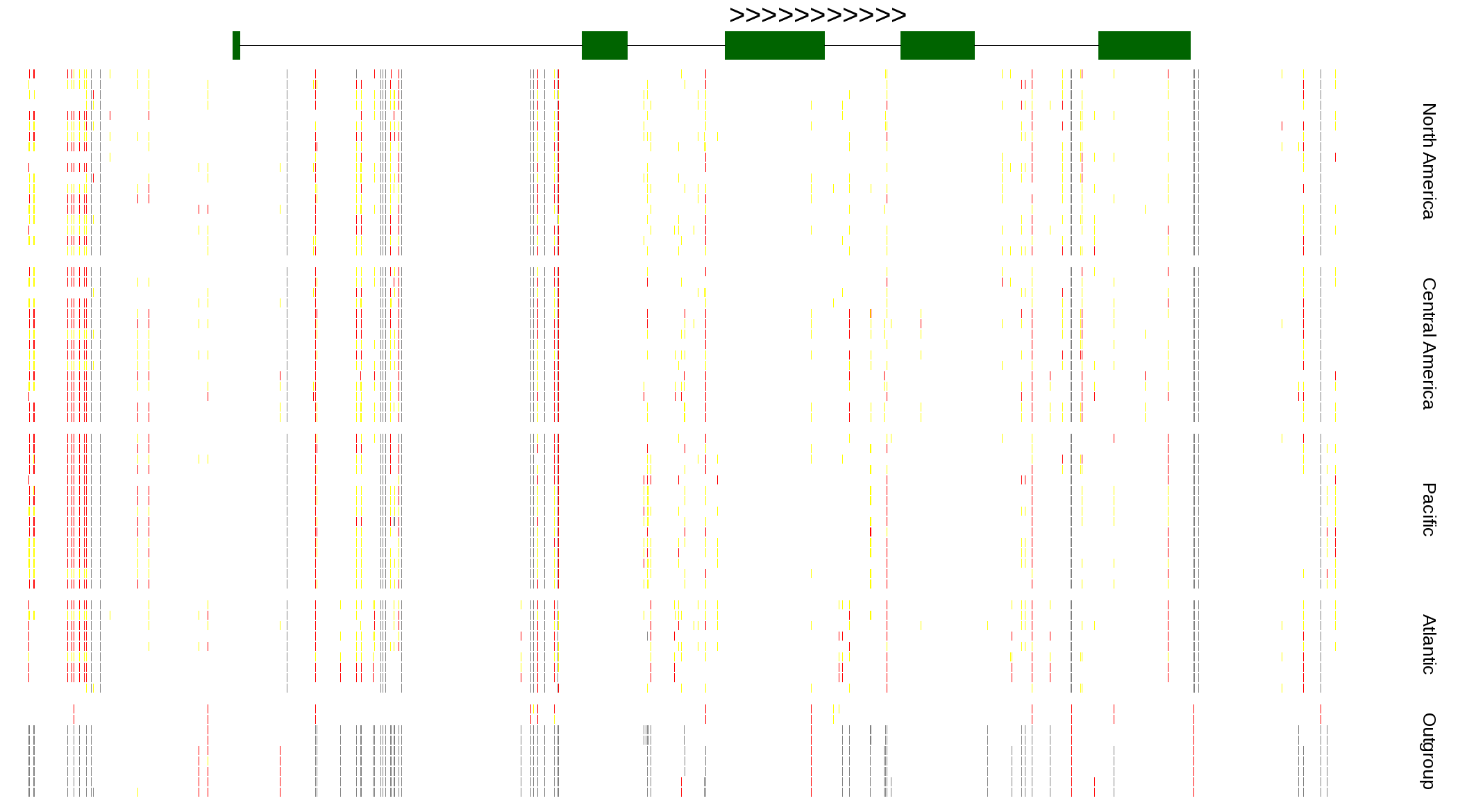
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS213607-TA
ATGGCTGGATTCAGCTGGTATCTCTCTGAAGGCTTCCAGGTAATTATTGAGAAGTCTAAAGAAGCGAAATGTTCCTTAAAAGATGTACAATTGCGTTTCTTGTGCCCTGATGACTTAGAAGAGGTCCGATGTCTCTGCAGGGATTGGTTTCCAATAGAATATCCACAATCATGGTATGAAGACATAACTTCTTCGGAGAGATTTTTTGCATTAGCAGCAGTTTATAAAACGCAGATTATTGGCTTAATAGTTGCAGAGATAAAACCTTATTTAAAATTAAACGCAGAAGACCGCGGCATATTGTCCCGATGGTTTGCATCAAAGGATACTCTTGTTGCATACATTCTTTCATTAGGTGTAGTCCGGGCATATCGTCGTTCAGGGGTAGCGACGATGCTTCTGGATGTACTCATCAATCACCTCTCAGGACCCATACCACAGCCGCCTCATGAGTATCGAGTCAAAGCCATATTTCTGCATGTACTCACTACTAACAATGAAGCCATACTTTTTTATGAAAAGAGAAGATTCAAACTGCACTCATTCCTGCCATATTACTACTCGATAAAGGGTAGATGTAAAGACGGGTTCACATATGTGTACTATGTGAACGGCGGCCACGCGCCCTGGGGTCTGTATGATTACGTGAAGTACGTGGCGCGGACCGCATGGAGAGGGGGCGGGCTCTACCCCTGGCTGTGGGGTAAGTTGACCGCCCTCACCATAGCCTGGCACAGGTAG
>DPOGS213607-PA
MAGFSWYLSEGFQVIIEKSKEAKCSLKDVQLRFLCPDDLEEVRCLCRDWFPIEYPQSWYEDITSSERFFALAAVYKTQIIGLIVAEIKPYLKLNAEDRGILSRWFASKDTLVAYILSLGVVRAYRRSGVATMLLDVLINHLSGPIPQPPHEYRVKAIFLHVLTTNNEAILFYEKRRFKLHSFLPYYYSIKGRCKDGFTYVYYVNGGHAPWGLYDYVKYVARTAWRGGGLYPWLWGKLTALTIAWHR-