| DPOGS213886 | ||
|---|---|---|
| Transcript | DPOGS213886-TA | 621 bp |
| Protein | DPOGS213886-PA | 206 aa |
| Genomic position | DPSCF300141 + 191160-192911 | |
| RNAseq coverage | 351x (Rank: top 33%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL006670 | 5e-71 | 89.61% | |
| Bombyx | BGIBMGA002559-TA | 2e-22 | 33.81% | |
| Drosophila | CG8188-PA | 1e-59 | 51.87% | |
| EBI UniRef50 | UniRef50_A7SE05 | 8e-57 | 50.48% | Ubiquitin-conjugating enzyme E2 S n=31 Tax=Eukaryota RepID=UBE2S_NEMVE |
| NCBI RefSeq | NP_001161423.1 | 8e-60 | 51.21% | hypothetical protein LOC100167236 [Acyrthosiphon pisum] |
| NCBI nr blastp | gi|269784617 | 1e-58 | 51.21% | ubiquitin-conjugating enzyme E2 S-like [Acyrthosiphon pisum] |
| NCBI nr blastx | gi|48095134 | 9e-58 | 52.36% | PREDICTED: ubiquitin-conjugating enzyme E2 S-like [Apis mellifera] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0016881 | 1e-33 | acid-amino acid ligase activity | |
| KEGG pathway | api:100167011 | 2e-59 | ||
| K10583 (UBE2S, E2EPF) | maps-> | Ubiquitin mediated proteolysis | ||
| InterPro domain | [10-160] IPR016135 | 3.6e-46 | Ubiquitin-conjugating enzyme/RWD-like | |
| [15-150] IPR000608 | 1e-33 | Ubiquitin-conjugating enzyme, E2 | ||
| Orthology group | MCL16627 | Patchy | ||
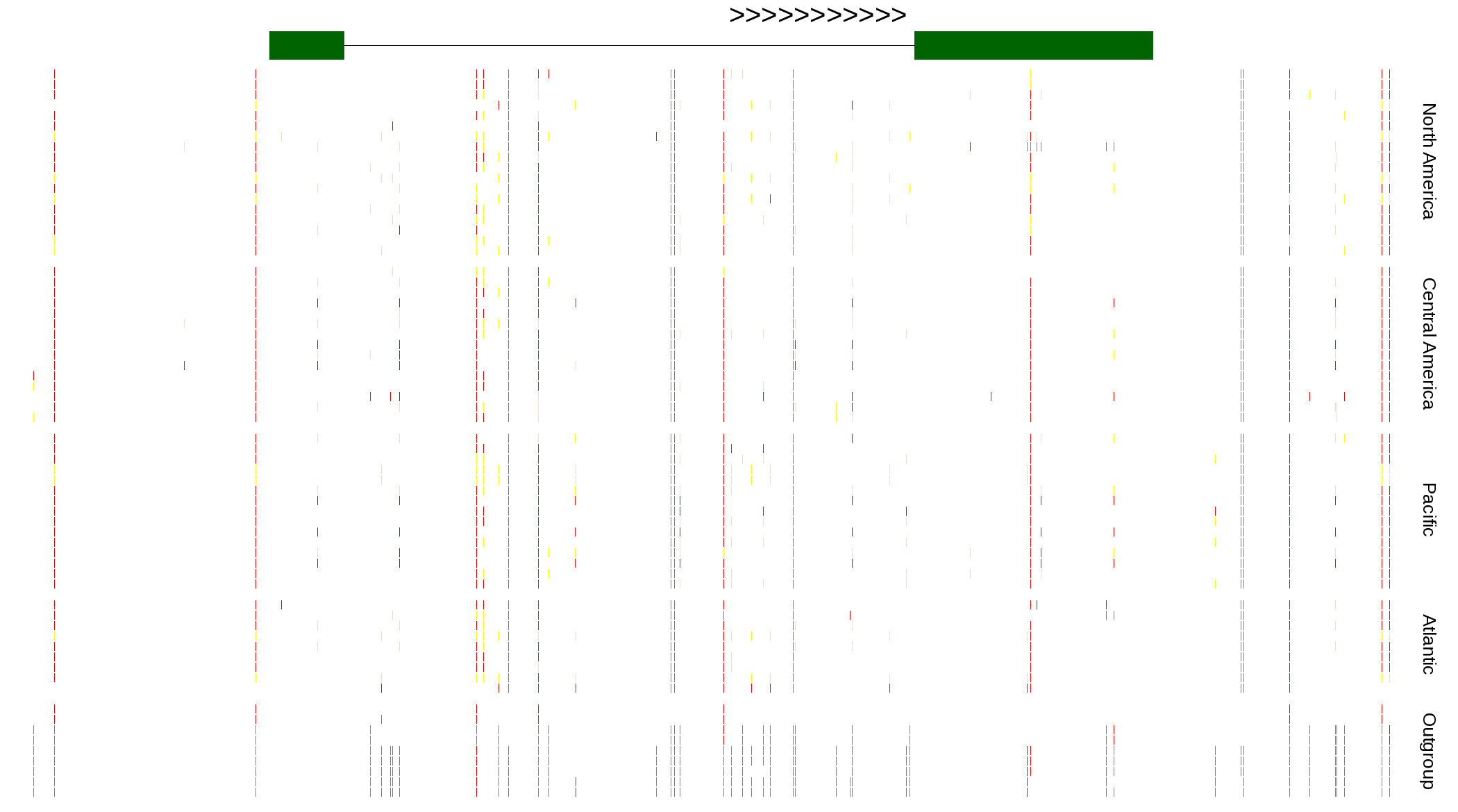
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS213886-TA
ATGTCAAACGTGGAGAACGTTTGCCCCCAAGTTCTTCGCGGAGTGACGAAGGAGCTACGGCGGCTGGCAGCTAATCCTCCAGCCGGCATCAAGCTGGTGCTGCGTGATGATGATATCACAGACGTGGTGGCGCTCATCGACGGCCCCGCGGACACGCCGTACGCTGGTGGCGTGTTCCGGGTGCGCCTGTGTCTGGGGCGCGAGTTCCCCACGGCCCCTCCTCGCGCGTTCTTCCTCACTCGCATCTTCCACCCCAACGTGTCGTCGGCGGGTGAGGTGTGCGTCAACACCTTGAAGCGCGATTGGCGGCCAGAGCTCGGACTGGAGCACGCTCTGCTCGCCGTCAAGTGTCTGCTGATCGCCCCCAACGCAGACTCGGCGCTCAACGCGGAGGCGGCCGCCTTACTCAGAGACCGCTACGACGACTACTTCGCCCGCGCCAAGCTCTACACCGACATACATGCGGGGGTCGGGGTGGCCGCGGTGTTTCGTGAGGAGGCGGCTGCGGCTGCGGCTGCGGCTGCGGAGGGGGACGGGGAAGGGGGGCCGAGGGTCAAGCGTGAGCGGCGCGTGGGAGCCCCGCCCGCAAGAGACAAGAGACGTATACTGAAGAGGTTATGA
>DPOGS213886-PA
MSNVENVCPQVLRGVTKELRRLAANPPAGIKLVLRDDDITDVVALIDGPADTPYAGGVFRVRLCLGREFPTAPPRAFFLTRIFHPNVSSAGEVCVNTLKRDWRPELGLEHALLAVKCLLIAPNADSALNAEAAALLRDRYDDYFARAKLYTDIHAGVGVAAVFREEAAAAAAAAAEGDGEGGPRVKRERRVGAPPARDKRRILKRL-