| DPOGS214108 | ||
|---|---|---|
| Transcript | DPOGS214108-TA | 579 bp |
| Protein | DPOGS214108-PA | 192 aa |
| Genomic position | DPSCF300014 - 1926844-1927422 | |
| RNAseq coverage | 62x (Rank: top 68%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL011410 | 5e-89 | 79.58% | |
| Bombyx | BGIBMGA006159-TA | 2e-77 | 70.16% | |
| Drosophila | CG8891-PA | 1e-69 | 64.48% | |
| EBI UniRef50 | UniRef50_Q9VMW7 | 2e-67 | 64.48% | Inosine triphosphate pyrophosphatase n=163 Tax=root RepID=ITPA_DROME |
| NCBI RefSeq | NP_001040500.1 | 5e-76 | 70.16% | inosine triphosphatase [Bombyx mori] |
| NCBI nr blastp | gi|114052993 | 8e-75 | 70.16% | inosine triphosphatase [Bombyx mori] |
| NCBI nr blastx | gi|157119275 | 2e-75 | 71.12% | inosine triphosphate pyrophosphatase (itpase) (inosine triphosphatase) [Aedes aegypti] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0016787 | 1e-108 | hydrolase activity | |
| KEGG pathway | aag:AaeL_AAEL008605 | 6e-75 | ||
| K01519 (ITPA) | maps-> | Purine metabolism | ||
| Drug metabolism - other enzymes | ||||
| Pyrimidine metabolism | ||||
| InterPro domain | [1-191] IPR002637 | 1e-108 | Ham1-like protein | |
| Orthology group | MCL13618 | Single-copy universal gene | ||
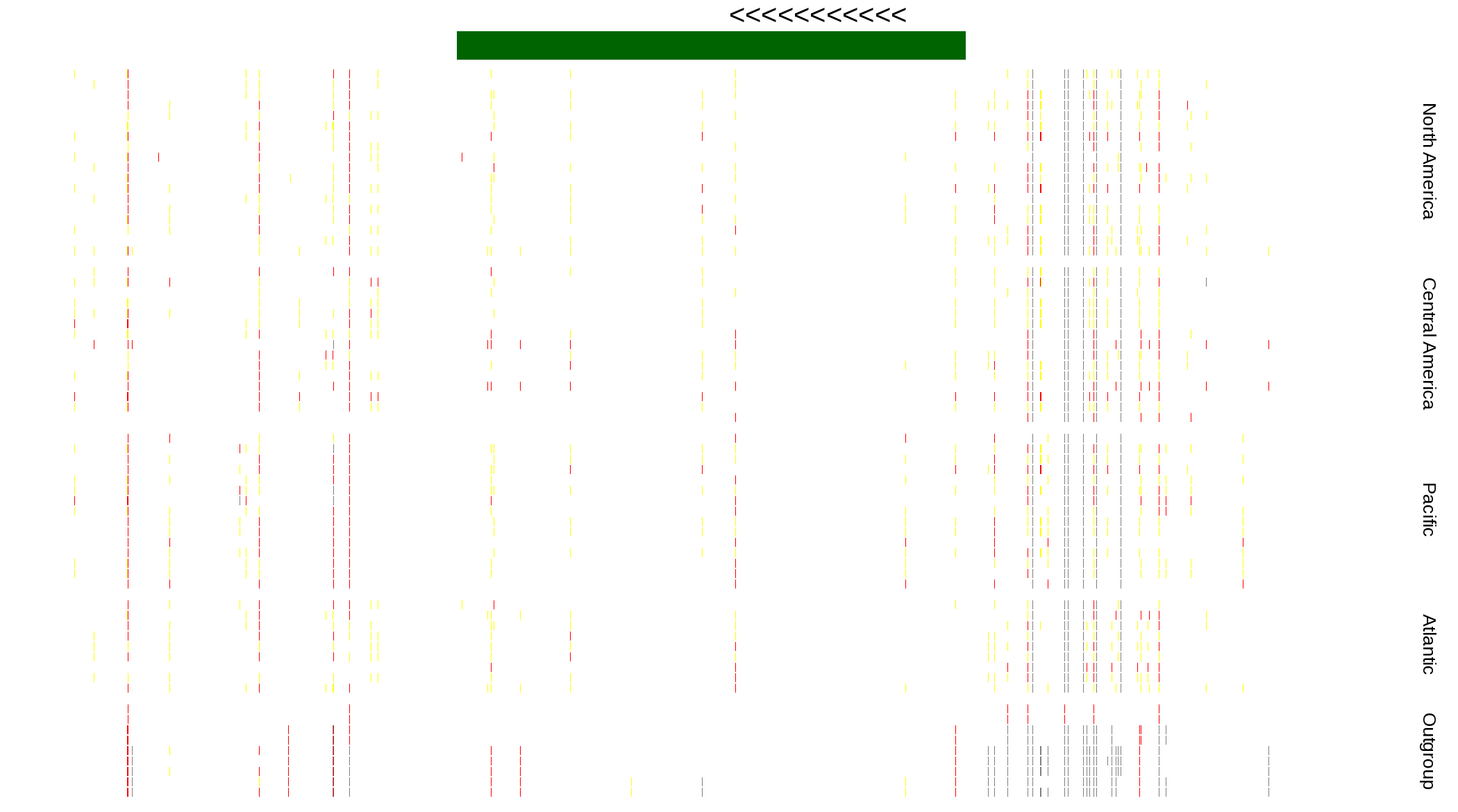
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS214108-TA
ATGAATCAAAGAGCTGTAGTTTTCGTCACTGGTAATTTAAAAAAATTGGAAGAGGTGAAATCTATTTTGGGTCATAACTTTCCATTAGAAGTTACAAATTATAGGTTAGATTTGCCTGAACTACAGGGAGAGTTGAATGAGGTATCTATCAGAAAATGTCAAGAAGCAGCTCGACGTTTAAAAAAGCCGGTATTTATAGAAGACACTTGCCTTTGTTTTAATGCATTGGAAGGTTTGCCTGGACCTTACATTAAATGGTTTTTAGAAAAGTTAAAACCAGAAGGTCTGTATAAATTACTTGAAGGCTGGACAGATAAATCTGCAGAAGCAGTATGTACATTCGCCTATAGTCCAGGAAGTGATAAAGAATCAGACATTGTATTATTTCAAGGAATAACAAAAGGAACAATTGTACCTCCAAGAGGTTCTAGAGATTTTGGATGGGATTGTATTTTCCAACCACTTGGTTATGACAAAACTTATGGCGAACTTCCTAAAGAAGAAAAAAACAAAATATCACATAGGTACAGAGCTCTTGATAAGTTGCGTGATTATTTTATAGTAAAAAACAATTGTTAG
>DPOGS214108-PA
MNQRAVVFVTGNLKKLEEVKSILGHNFPLEVTNYRLDLPELQGELNEVSIRKCQEAARRLKKPVFIEDTCLCFNALEGLPGPYIKWFLEKLKPEGLYKLLEGWTDKSAEAVCTFAYSPGSDKESDIVLFQGITKGTIVPPRGSRDFGWDCIFQPLGYDKTYGELPKEEKNKISHRYRALDKLRDYFIVKNNC-