| DPOGS214943 | ||
|---|---|---|
| Transcript | DPOGS214943-TA | 762 bp |
| Protein | DPOGS214943-PA | 253 aa |
| Genomic position | DPSCF300280 - 105195-107210 | |
| RNAseq coverage | 2405x (Rank: top 5%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL015591 | 9e-104 | 82.61% | |
| Bombyx | BGIBMGA004824-TA | 1e-132 | 93.68% | |
| Drosophila | CG7834-PB | 7e-117 | 81.82% | |
| EBI UniRef50 | UniRef50_Q9DCW4 | 5e-99 | 70.52% | Electron transfer flavoprotein subunit beta n=120 Tax=cellular organisms RepID=ETFB_MOUSE |
| NCBI RefSeq | NP_001040123.1 | 4e-131 | 93.68% | electron-transfer-flavoprotein beta polypeptide [Bombyx mori] |
| NCBI nr blastp | gi|114053151 | 7e-130 | 93.68% | electron-transfer-flavoprotein beta polypeptide [Bombyx mori] |
| NCBI nr blastx | gi|114053151 | 1e-130 | 93.68% | electron-transfer-flavoprotein beta polypeptide [Bombyx mori] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0009055 | 6.7e-170 | electron carrier activity | |
| KEGG pathway | ||||
| InterPro domain | [1-254] IPR012255 | 6.7e-170 | Electron transfer flavoprotein, beta subunit | |
| [3-249] IPR014729 | 3e-106 | Rossmann-like alpha/beta/alpha sandwich fold | ||
| [24-216] IPR014730 | 1.2e-70 | Electron transfer flavoprotein, alpha/beta-subunit, N-terminal | ||
| Orthology group | MCL13492 | Single-copy universal gene | ||
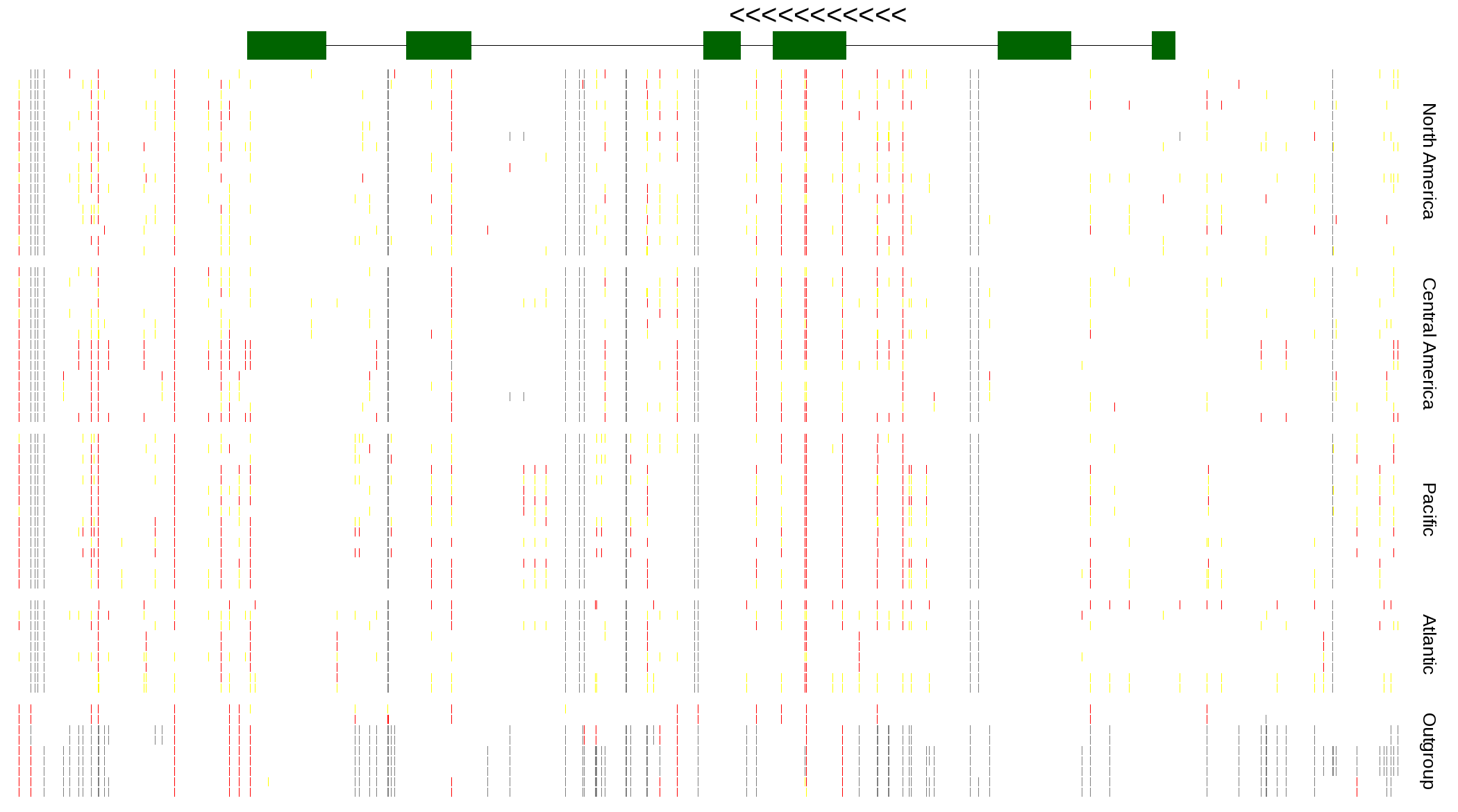
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS214943-TA
ATGGCTCGTGTATTAGTAGGTGTTAAGAGAGTTATTGATTATGCAGTCAAGATTCGTGTAAAGCCCGACAAGTCAGGAGTAGTCACGGATGGGGTGAAACATTCAATGAACCCTTTTGATGAGATTGCAGTGGAAGAAGCTGTTAGGATGAAGGAGAAAAAGTTAGCTAGTGAAGTCATCGCAGTATCCTGTGGTCCTACTCAAGCACAGGAAACTCTGAGAACAGCTCTAGCTATGGGTGCTGATCGAGCTATCCATGTTGAAGTTGCTGGAGCTGAATATGAAACCTTACAGCCACTCCATGTGGCAAAGATTCTAGCTAAACTCTCACAGGATGAGAAGGCTGATCTTGTGATAGTGGGAAAACAGGCAATTGATGATGACTCAAACCAAACTGCACAAATGACGGCCGCCATTCTGGATTGGCCTCAAGGAACATTTGCTTCCAAGGTGGAGAAGAGTGACTCTGGTTTGACTGTGACTCGTGAGATTGATGGTGGTCTGGAGGTCATTAAGACCAAGCTGCCAGCCGTCATCAGCGCGGATCTTCGTCTCAACGAGCCCAGATATGCCACTCTACCAAATATTATGAAAGCAAAAAAGAAGCCAATGAAGAAAGTATCAGCAAAGGATCTCGGTGTGGACTTAGCTCCCAGGATAAAGATACTGAGTGTTGAAGACCCTCCAGTGAGACAGGCTGGGTCTATTGTCCCGGATGTGGACACACTCGTCGCCAAACTCAAGGAAGGGGGCCATGTTTAA
>DPOGS214943-PA
MARVLVGVKRVIDYAVKIRVKPDKSGVVTDGVKHSMNPFDEIAVEEAVRMKEKKLASEVIAVSCGPTQAQETLRTALAMGADRAIHVEVAGAEYETLQPLHVAKILAKLSQDEKADLVIVGKQAIDDDSNQTAQMTAAILDWPQGTFASKVEKSDSGLTVTREIDGGLEVIKTKLPAVISADLRLNEPRYATLPNIMKAKKKPMKKVSAKDLGVDLAPRIKILSVEDPPVRQAGSIVPDVDTLVAKLKEGGHV-