| DPOGS215238 | ||
|---|---|---|
| Transcript | DPOGS215238-TA | 507 bp |
| Protein | DPOGS215238-PA | 168 aa |
| Genomic position | DPSCF300047 - 832747-839803 | |
| RNAseq coverage | 1102x (Rank: top 11%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL020584 | 5e-73 | 87.43% | |
| Bombyx | BGIBMGA001512-TA | 2e-60 | 84.73% | |
| Drosophila | gish-PJ | 1e-54 | 61.85% | |
| EBI UniRef50 | UniRef50_UPI0002246AE1 | 1e-60 | 73.17% | UPI0002246AE1 related cluster n=1 Tax=unknown RepID=UPI0002246AE1 |
| NCBI RefSeq | XP_001607046.1 | 1e-59 | 72.56% | PREDICTED: similar to CG6963-PE [Nasonia vitripennis] |
| NCBI nr blastp | gi|345485756 | 4e-60 | 73.17% | PREDICTED: casein kinase I isoform gamma-3-like [Nasonia vitripennis] |
| NCBI nr blastx | gi|347971044 | 5e-61 | 80.69% | AGAP003997-PB [Anopheles gambiae str. PEST] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0016772 | 1.8e-26 | transferase activity, transferring phosphorus-containing groups | |
| GO:0005524 | 9.2e-07 | ATP binding | ||
| GO:0004672 | 9.2e-07 | protein kinase activity | ||
| GO:0006468 | 9.2e-07 | protein phosphorylation | ||
| KEGG pathway | nvi:100123405 | 3e-59 | ||
| K08958 (CSNK1G) | maps-> | Hedgehog signaling pathway | ||
| InterPro domain | [52-166] IPR011009 | 1.8e-26 | Protein kinase-like domain | |
| [57-163] IPR017442 | 9.2e-07 | Serine/threonine-protein kinase-like domain | ||
| Orthology group | MCL35053 | Lepidoptera specific | ||
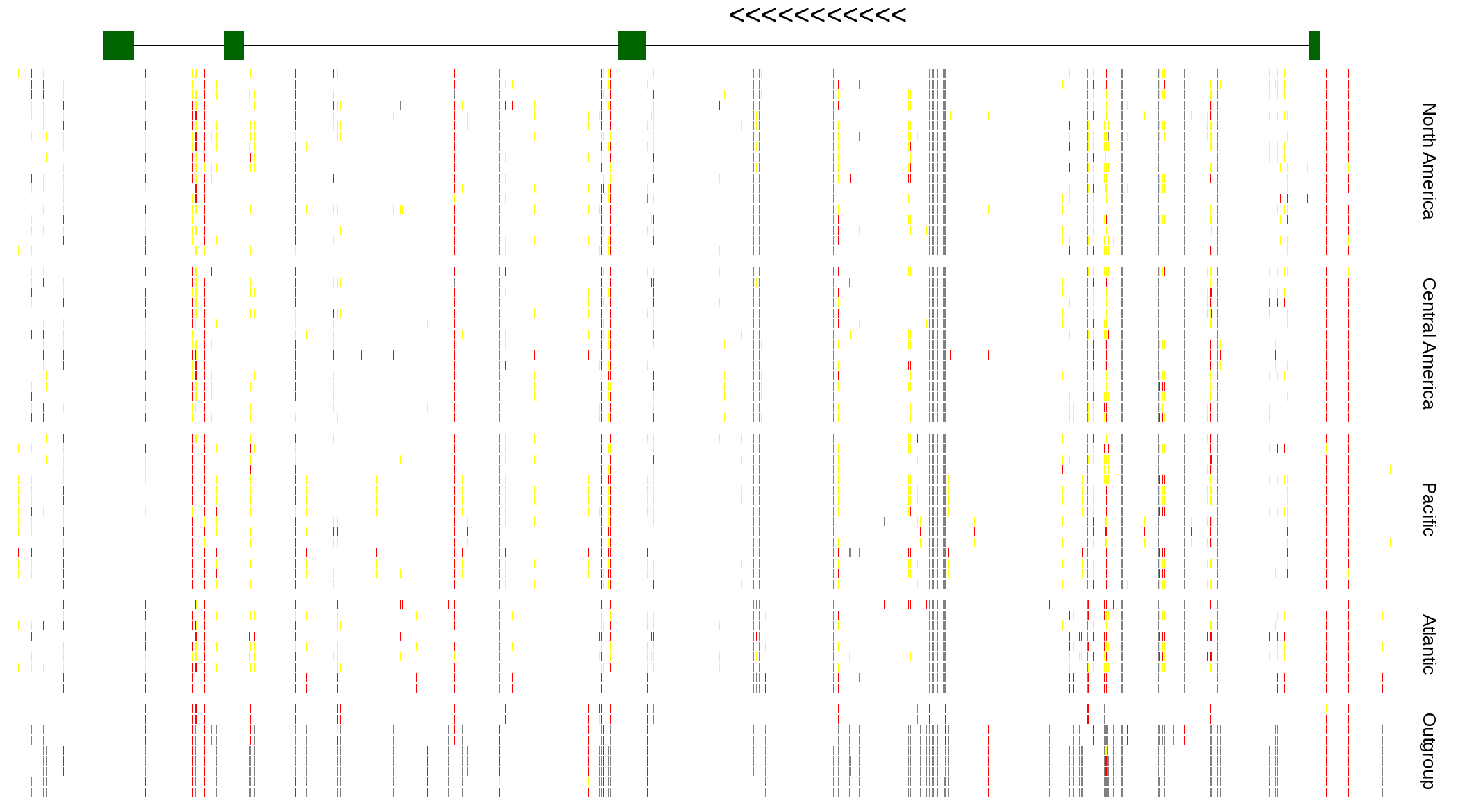
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS215238-TA
ATGAATAAACGTGAGGGTACCAAGGACAAGGAACGTCAGAGCGACGACAAAAGAACTTCCACCGTAACCGGTGCGGTGGGCGGGCGCGGCGTGGGCGGGGGCAGCACGATGCACACGTCGCGGCACTCGGTCACCTCGTCCTCGGGCGTCCTTATGGTGGGGCCAAACTTCCGCGTCGGCAAGAAAATTGGCTGCGGGAACTTCGGCGAGCTTAGGCTCGGGAAAAATCTATACAACAATGAACACGTAGCTATTAAGATGGAACCTATGAAGTCGAAGGCTCCACAACTACATCTAGAGTACAGATTTTACAAGTTATTAGGAACCCACGAGGGAATACCAAAAGTATACTACTTGGGAACATGCGGTGGTCGCTACAACGCCATGGTCATGGAGTTGCTAGGACCCTCATTAGAGGATCTCTTCGTACACTGCGGTAGGAGATTCAGACTGAAGACTGTACTCCTGATCGCTATACAGCTTGTAAGTTGGTTGAACGAAAAGTAG
>DPOGS215238-PA
MNKREGTKDKERQSDDKRTSTVTGAVGGRGVGGGSTMHTSRHSVTSSSGVLMVGPNFRVGKKIGCGNFGELRLGKNLYNNEHVAIKMEPMKSKAPQLHLEYRFYKLLGTHEGIPKVYYLGTCGGRYNAMVMELLGPSLEDLFVHCGRRFRLKTVLLIAIQLVSWLNEK-