| DPOGS215422 | ||
|---|---|---|
| Transcript | DPOGS215422-TA | 351 bp |
| Protein | DPOGS215422-PA | 116 aa |
| Genomic position | DPSCF300088 + 1217849-1219446 | |
| RNAseq coverage | 0x (Rank: top 99%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL003453 | 2e-21 | 62.86% | |
| Bombyx | BGIBMGA012386-TA | 2e-21 | 65.22% | |
| Drosophila | Cyp6d5-PA | 3e-08 | 36.23% | |
| EBI UniRef50 | UniRef50_D5L0N0 | 1e-12 | 53.25% | Cytochrome P450 6AN5 n=7 Tax=Obtectomera RepID=D5L0N0_MANSE |
| NCBI RefSeq | NP_001073135.1 | 1e-19 | 65.22% | cytochrome P450 6AB4 [Bombyx mori] |
| NCBI nr blastp | gi|116292895 | 9e-19 | 66.67% | CYP6AB4 [Bombyx mandarina] |
| NCBI nr blastx | gi|116292895 | 3e-18 | 66.67% | CYP6AB4 [Bombyx mandarina] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0009055 | 3.2e-13 | electron carrier activity | |
| GO:0020037 | 3.2e-13 | heme binding | ||
| GO:0016705 | 3.2e-13 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen | ||
| GO:0005506 | 3.2e-13 | iron ion binding | ||
| GO:0055114 | 3.2e-13 | oxidation-reduction process | ||
| GO:0004497 | 1.3e-05 | monooxygenase activity | ||
| KEGG pathway | phu:Phum_PHUM417190 | 2e-08 | ||
| K07424 (CYP3A) | maps-> | Drug metabolism - cytochrome P450 | ||
| Drug metabolism - other enzymes | ||||
| Linoleic acid metabolism | ||||
| Steroid hormone biosynthesis | ||||
| Metabolism of xenobiotics by cytochrome P450 | ||||
| gamma-Hexachlorocyclohexane degradation | ||||
| Retinol metabolism | ||||
| InterPro domain | [1-64] IPR001128 | 3.2e-13 | Cytochrome P450 | |
| Orthology group | ||||
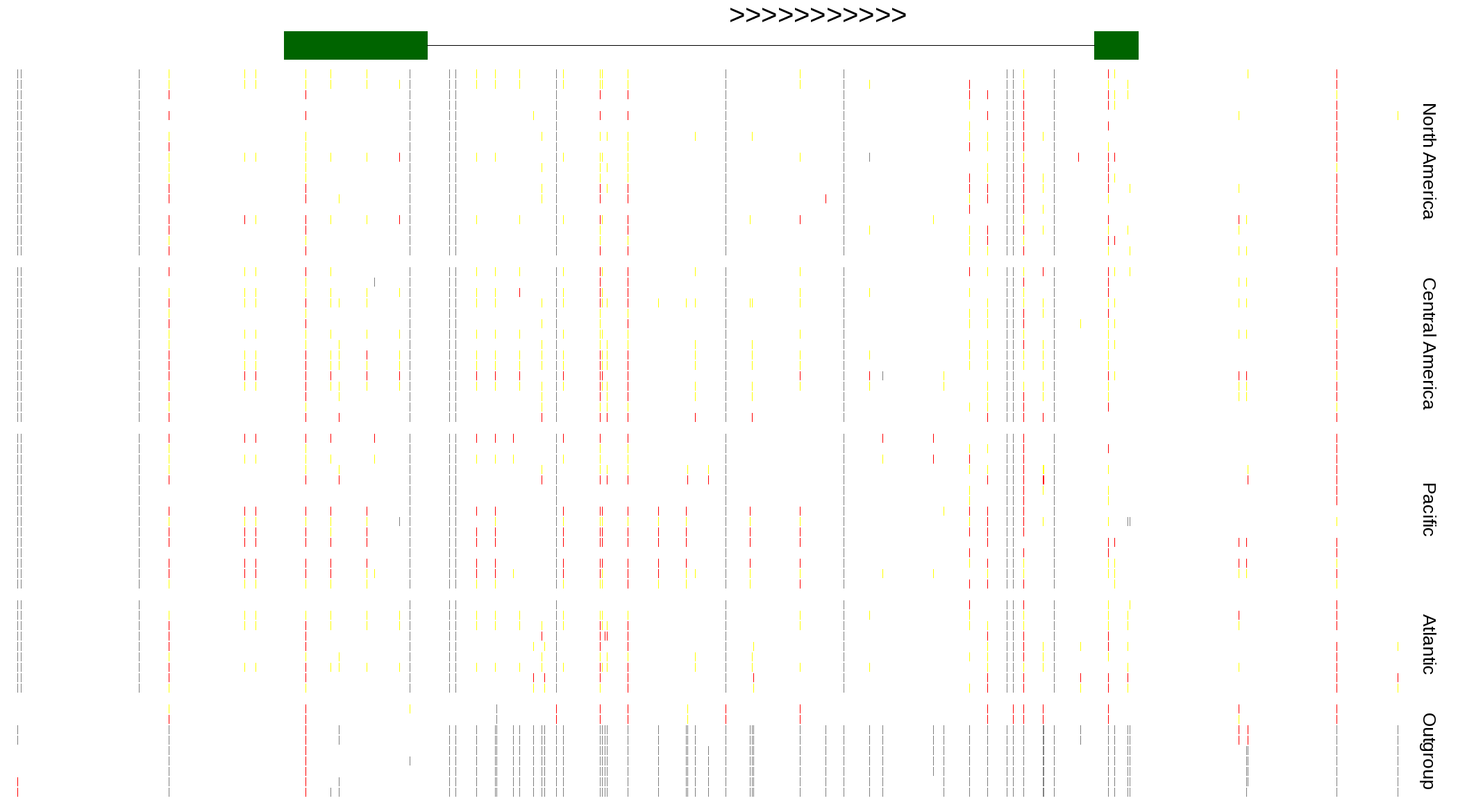
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS215422-TA
ATGACCTATTTGGAGTCGGCTTTCAAAGAGGGGATGAGAACCTTTCCATCCCTAGGGTTTCTTGTAAGACAATGCGCCAAACGCTACACATTCGAAGATCTCATAATCACACTTCAAGCGCTGCACAACGATCCGAAATACTTCGAAAACCCGACCGAATTTCGCCCGGAAAGGTTCACCCGACCATCTCCAGCGGCCATGATAAGTTCGTGTACCTGCCGTTTGGCGAGGGTCCGCGCGCGTGTGTCGGCGAGCGGCTGGGTAATAGGAAACTTAGCAGTCGGGCGTTTCAATCAACTTATACATAAGACATTCCATGACTGCGCCACAGACGACCGCTACTACTGCTAG
>DPOGS215422-PA
MTYLESAFKEGMRTFPSLGFLVRQCAKRYTFEDLIITLQALHNDPKYFENPTEFRPERFTRPSPAAMISSCTCRLARVRARVSASGWVIGNLAVGRFNQLIHKTFHDCATDDRYYC-