| DPOGS215517 | ||
|---|---|---|
| Transcript | DPOGS215517-TA | 732 bp |
| Protein | DPOGS215517-PA | 243 aa |
| Genomic position | DPSCF300467 - 91902-94726 | |
| RNAseq coverage | 267x (Rank: top 40%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL005536 | 1e-117 | 92.17% | |
| Bombyx | BGIBMGA004224-TA | 1e-67 | 91.41% | |
| Drosophila | ras-PC | 2e-21 | 29.66% | |
| EBI UniRef50 | UniRef50_Q9P2T1 | 5e-105 | 74.07% | GMP reductase 2 n=638 Tax=root RepID=GMPR2_HUMAN |
| NCBI RefSeq | XP_001603068.1 | 2e-116 | 82.64% | PREDICTED: similar to Gmpr2 protein [Nasonia vitripennis] |
| NCBI nr blastp | gi|156547587 | 3e-115 | 82.64% | PREDICTED: GMP reductase 1-like [Nasonia vitripennis] |
| NCBI nr blastx | gi|156547587 | 8e-114 | 82.64% | PREDICTED: GMP reductase 1-like [Nasonia vitripennis] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0008152 | 6.2e-94 | metabolic process | |
| GO:0003824 | 6.2e-94 | catalytic activity | ||
| GO:0055114 | 5.3e-89 | oxidation-reduction process | ||
| KEGG pathway | nvi:100119268 | 6e-116 | ||
| K00364 (E1.7.1.7, guaC) | maps-> | Purine metabolism | ||
| InterPro domain | [6-242] IPR013785 | 6.2e-94 | Aldolase-type TIM barrel | |
| [1-237] IPR001093 | 5.3e-89 | IMP dehydrogenase/GMP reductase | ||
| Orthology group | MCL16929 | Patchy | ||
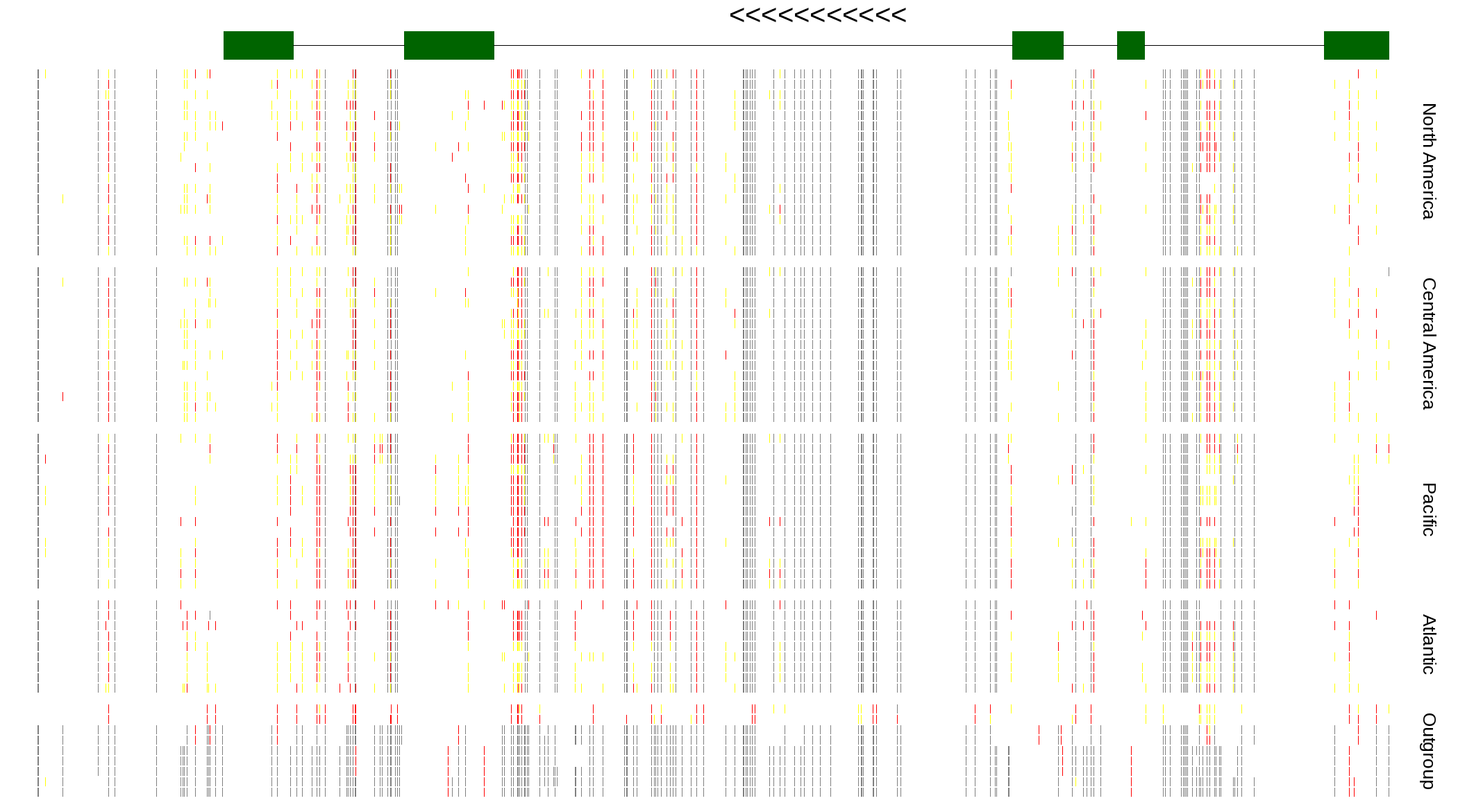
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS215517-TA
GGTACCGCGGAGGCTGACTTCGACCGTCTATCAGAGATCTTGAACAGCGTCAGTAGTTTGAAATTCATTTGTCTCGACGTCGCCAACGGATACTCGCAGCACTTCGTGGAGTACGTGAGGCGAGTGAGGGCGGCATTCCCATCACACACCATCATTGCTGGTAACGTGGTGACTGGGGAGATGGTGGAGGAGCTCATCCTGTCAGGAGCTGATATCATAAAGGTCGGTATCGGTCCGGGGTCGGTGTGCACGACCCGCATGAAGACGGGCGTGGGATACCCTCAGCTGTCCGCCGTCATAGAGTGCGCGGACGCCGCCCATGGGCTCAAGGGACACATTATATCTGACGGCGGTTGTACTTGTCCTGGCGACGTGGCGAAAGCCTTCGGAGCCGGCGCCGACTTCGTGATGGCCGGCGGCATGTTCGCGGGACACGACCAGTGCTCCGGGGACCTCGTCACCAAACCCGATGGAAAGAAAGTGAAACTGTTCTATGGAATGTCCTCATCGACCGCCATGTTGAAACACTCGGGCGGCGTCGCTGAATACAGATCCTCTGAAGGTAAAACGGTGGAAGTTGAATACCGAGGCAACGTAGAGGTCACGGTGAAGGACGTCCTGGGCGGCTTGAGGTCGGCGTGCACGTACGTGGGGGCCTCGAGGCTCAGGGAACTGCCTCGGAGGGCGACCTTCATCAGGTGTTGCAGACAAGTCAACGACGTCTTCTCTTAA
>DPOGS215517-PA
GTAEADFDRLSEILNSVSSLKFICLDVANGYSQHFVEYVRRVRAAFPSHTIIAGNVVTGEMVEELILSGADIIKVGIGPGSVCTTRMKTGVGYPQLSAVIECADAAHGLKGHIISDGGCTCPGDVAKAFGAGADFVMAGGMFAGHDQCSGDLVTKPDGKKVKLFYGMSSSTAMLKHSGGVAEYRSSEGKTVEVEYRGNVEVTVKDVLGGLRSACTYVGASRLRELPRRATFIRCCRQVNDVFS-