| DPOGS215777 | ||
|---|---|---|
| Transcript | DPOGS215777-TA | 645 bp |
| Protein | DPOGS215777-PA | 214 aa |
| Genomic position | DPSCF300041 + 1807860-1810132 | |
| RNAseq coverage | 177x (Rank: top 50%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL014093 | 7e-42 | 46.99% | |
| Bombyx | BGIBMGA003537-TA | 4e-42 | 46.24% | |
| Drosophila | CG4306-PA | 1e-34 | 40.23% | |
| EBI UniRef50 | UniRef50_UPI0001A5CE84 | 5e-38 | 45.45% | UPI0001A5CE84 related cluster n=2 Tax=unknown RepID=UPI0001A5CE84 |
| NCBI RefSeq | NP_001155144.1 | 1e-38 | 45.45% | gamma-glutamyl cyclotransferase-like venom protein isoform 1 [Nasonia vitripennis] |
| NCBI nr blastp | gi|239735530 | 2e-37 | 45.45% | gamma-glutamyl cyclotransferase-like venom protein isoform 1 precursor [Nasonia vitripennis] |
| NCBI nr blastx | gi|239735530 | 7e-37 | 45.00% | gamma-glutamyl cyclotransferase-like venom protein isoform 1 precursor [Nasonia vitripennis] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0003839 | 3.7e-51 | gamma-glutamylcyclotransferase activity | |
| KEGG pathway | bta:533374 | 8e-28 | ||
| K00682 (GGCT) | maps-> | Glutathione metabolism | ||
| InterPro domain | [33-193] IPR017939 | 3.7e-51 | Gamma-glutamylcyclotransferase | |
| [30-194] IPR013024 | 4.8e-46 | Butirosin biosynthesis, BtrG-like | ||
| [86-143] IPR009288 | 8.4e-06 | AIG2-like | ||
| Orthology group | MCL10611 | Multiple-copy universal gene | ||
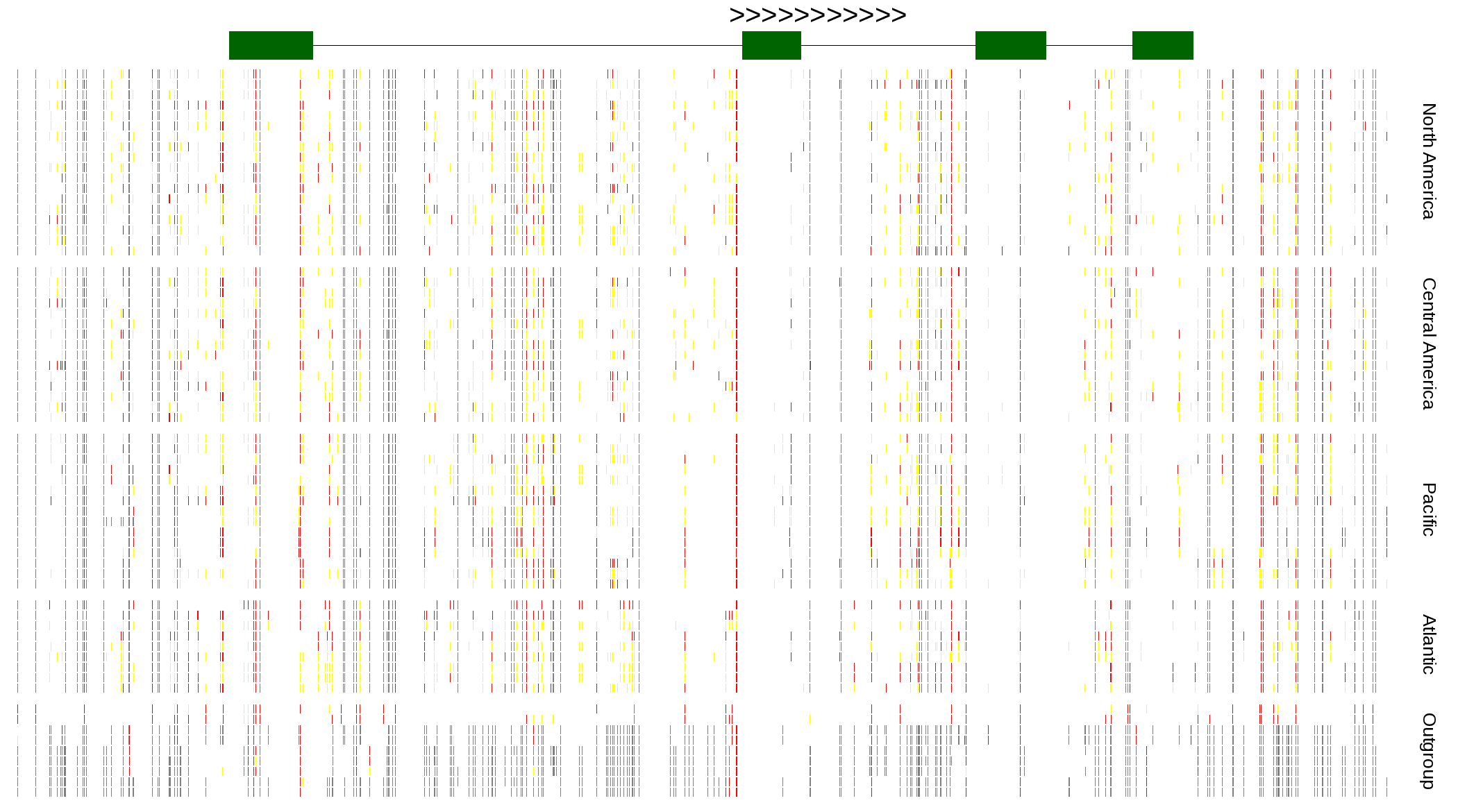
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS215777-TA
ATGGAGTCGTTCAAGATAGCAGTGTACATTTCTATACTGTTTTCGTCGCACATATTTGAACTAGTATTGACTAATGATGGAGTAGATTCAAAAAGTGAAAACAAATTTCTCTATTTCGCGTATGGAAGTAATCTCCTTGCGAGAAGAGTGCATATTAACAACCCAACAGCGGAGTTTTTTTCCTCTGCAAAGTTACCGAACTTTAGGTTAGACTTCCAAAGCTTCGACACGGAGTTTTGGAACGGTGCTGATGCGACCATCGTTGATGATTCGGACAGCGTTGTTTGGGGAGCGCTGTGGACACTTGACGTGAAAGATATACCTCATCTGGATGAGCAAGAGGGCATTCACCTTGGCCAGTACCGAACGAGGAACGTGTCGGTGGTAACTCCTGATGGTAGGTCAATGGTGGCTCGGACGTATCAGATGACACTAGAACCGCCTAAATTACCTGCTAAAGATATACCACCAGAACGACAACCTTCTGACACGTATATGGAGGTCATAATACGCGGCGCCTTTGAATGCTCGCTGCCTATGCATTACCTGCGGTTTCTTTTTACTTTTCCCACAAACGGTAAACTCGCTAGCTGGACTACAAGGCAACGTCTGGGTTACCCATTTAATGTAACTGAATTAAAATAA
>DPOGS215777-PA
MESFKIAVYISILFSSHIFELVLTNDGVDSKSENKFLYFAYGSNLLARRVHINNPTAEFFSSAKLPNFRLDFQSFDTEFWNGADATIVDDSDSVVWGALWTLDVKDIPHLDEQEGIHLGQYRTRNVSVVTPDGRSMVARTYQMTLEPPKLPAKDIPPERQPSDTYMEVIIRGAFECSLPMHYLRFLFTFPTNGKLASWTTRQRLGYPFNVTELK-