| DPOGS215816 | ||
|---|---|---|
| Transcript | DPOGS215816-TA | 534 bp |
| Protein | DPOGS215816-PA | 177 aa |
| Genomic position | DPSCF300073 - 447261-448279 | |
| RNAseq coverage | 149x (Rank: top 54%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL011648 | 1e-81 | 77.78% | |
| Bombyx | BGIBMGA013557-TA | 1e-50 | 74.79% | |
| Drosophila | ninaA-PA | 5e-45 | 44.38% | |
| EBI UniRef50 | UniRef50_Q7Q137 | 4e-58 | 59.66% | Peptidyl-prolyl cis-trans isomerase n=15 Tax=Endopterygota RepID=Q7Q137_ANOGA |
| NCBI RefSeq | XP_319131.4 | 9e-59 | 59.66% | AGAP009991-PA [Anopheles gambiae str. PEST] |
| NCBI nr blastp | gi|345483318 | 3e-59 | 59.89% | PREDICTED: peptidyl-prolyl cis-trans isomerase, rhodopsin-specific isozyme-like [Nasonia vitripennis] |
| NCBI nr blastx | gi|345483318 | 1e-59 | 59.89% | PREDICTED: peptidyl-prolyl cis-trans isomerase, rhodopsin-specific isozyme-like [Nasonia vitripennis] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0006457 | 2e-49 | protein folding | |
| GO:0003755 | 2e-49 | peptidyl-prolyl cis-trans isomerase activity | ||
| KEGG pathway | ||||
| InterPro domain | [2-132] IPR002130 | 2e-49 | Peptidyl-prolyl cis-trans isomerase, cyclophilin-type | |
| [2-133] IPR015891 | 8.3e-48 | Cyclophilin-like | ||
| Orthology group | MCL16896 | Insect specific | ||
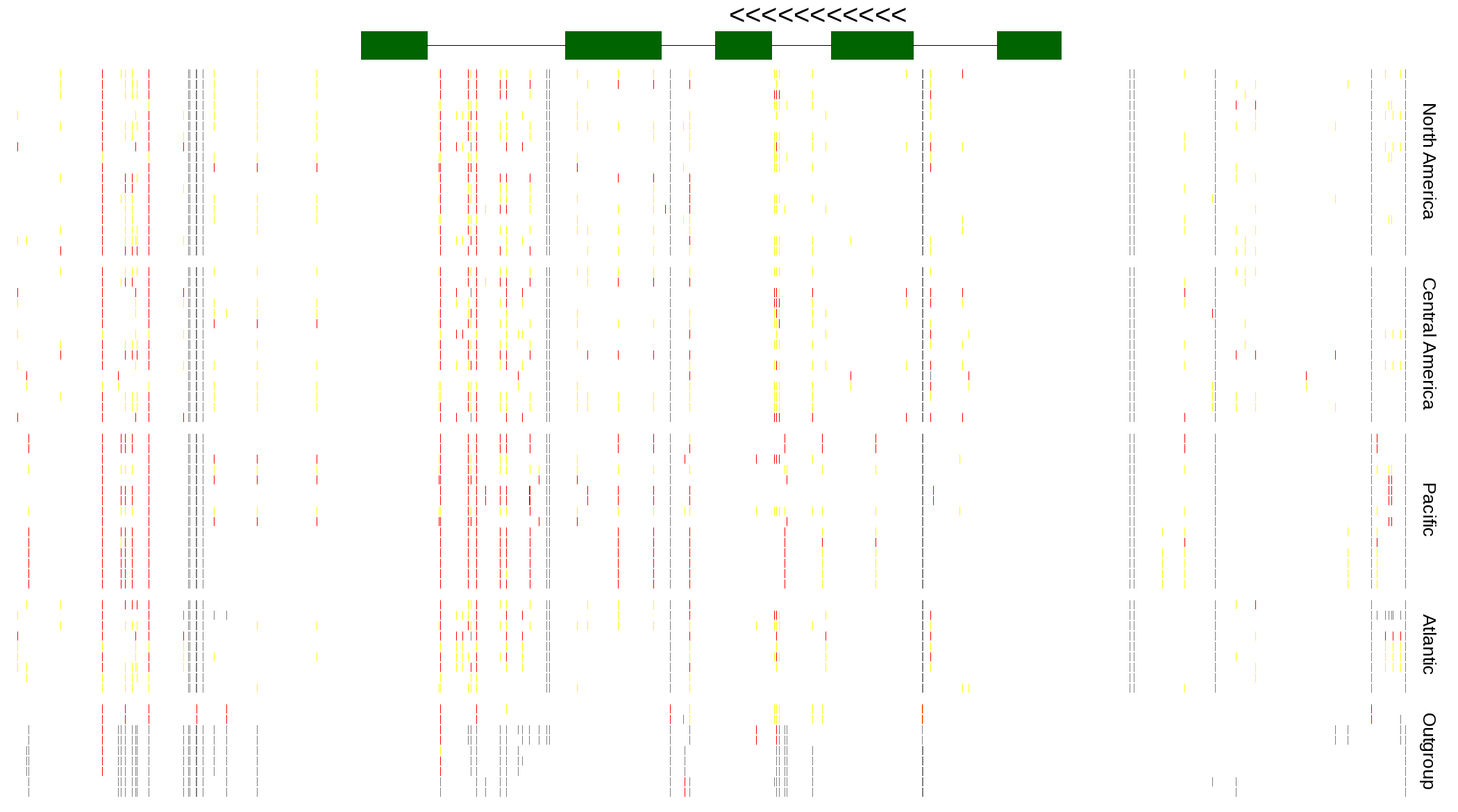
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS215816-TA
ATGAATTTCAAAGTACTTGCAACAAAAGGAATTAAAGGAAGATCATATAAAGGAACAAGTTTTAATCGTATAATAAAACGCTTTATGATACAAGGAGGTGATGTTGTATCAGACGATGGAACAGGTTCAATAAGTATATACGGTAAAACATTTAAAGATGAAAACTTAGAAACCCAACACACAGATGCTGGCTTTGTGTCAATGGCGAATAAAGGTAAGGATACAAATGGTTGTCAATTTATAATAACGACGAAACCAACACCTTGGCTTGATAACTTACATACGGTTGTCGGAAAAGTAGTGGAAGGTCAAAAGATAGTTCACATGTTAGAGCAGACTCCAACTGATATTAACGATCGGCCGACAGTGCGCGTGTACATCGTGGACTGCGGGCTGTTGTCCACTGAACCTTTTTATGTGTCTGATGAGCCCTATGACCTTTGGGGTTGGATCAAGGTGTCAGCCGCACCGCTGTCAATGTCATTCTCCATATTGGCTTTCTTTCATTGGATGATAAAAAAGATGGAGATTTAA
>DPOGS215816-PA
MNFKVLATKGIKGRSYKGTSFNRIIKRFMIQGGDVVSDDGTGSISIYGKTFKDENLETQHTDAGFVSMANKGKDTNGCQFIITTKPTPWLDNLHTVVGKVVEGQKIVHMLEQTPTDINDRPTVRVYIVDCGLLSTEPFYVSDEPYDLWGWIKVSAAPLSMSFSILAFFHWMIKKMEI-