| DPOGS200404 | ||
|---|---|---|
| Transcript | DPOGS200404-TA | 480 bp |
| Protein | DPOGS200404-PA | 159 aa |
| Genomic position | DPSCF300236 - 561053-563667 | |
| RNAseq coverage | 1741x (Rank: top 7%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | % | |||
| Bombyx | BGIBMGA008981-TA | 1e-60 | 72.46% | |
| Drosophila | Tak1-PA | 4e-11 | 38.64% | |
| EBI UniRef50 | UniRef50_E2AU44 | 7e-19 | 50.00% | Mitogen-activated protein kinase kinase kinase 7 n=8 Tax=Aculeata RepID=E2AU44_CAMFO |
| NCBI RefSeq | XP_397248.3 | 9e-19 | 42.98% | PREDICTED: similar to Mitogen-activated protein kinase kinase kinase 7 (Transforming growth factor-beta-activated kinase 1) (TGF-beta-activated kinase 1) [Apis mellifera] |
| NCBI nr blastp | gi|340713629 | 1e-22 | 52.88% | PREDICTED: mitogen-activated protein kinase kinase kinase 7 [Bombus terrestris] |
| NCBI nr blastx | gi|313907139 | 2e-23 | 51.82% | mitogen-activated protein kinase kinase kinase 7 [Apis cerana cerana] |
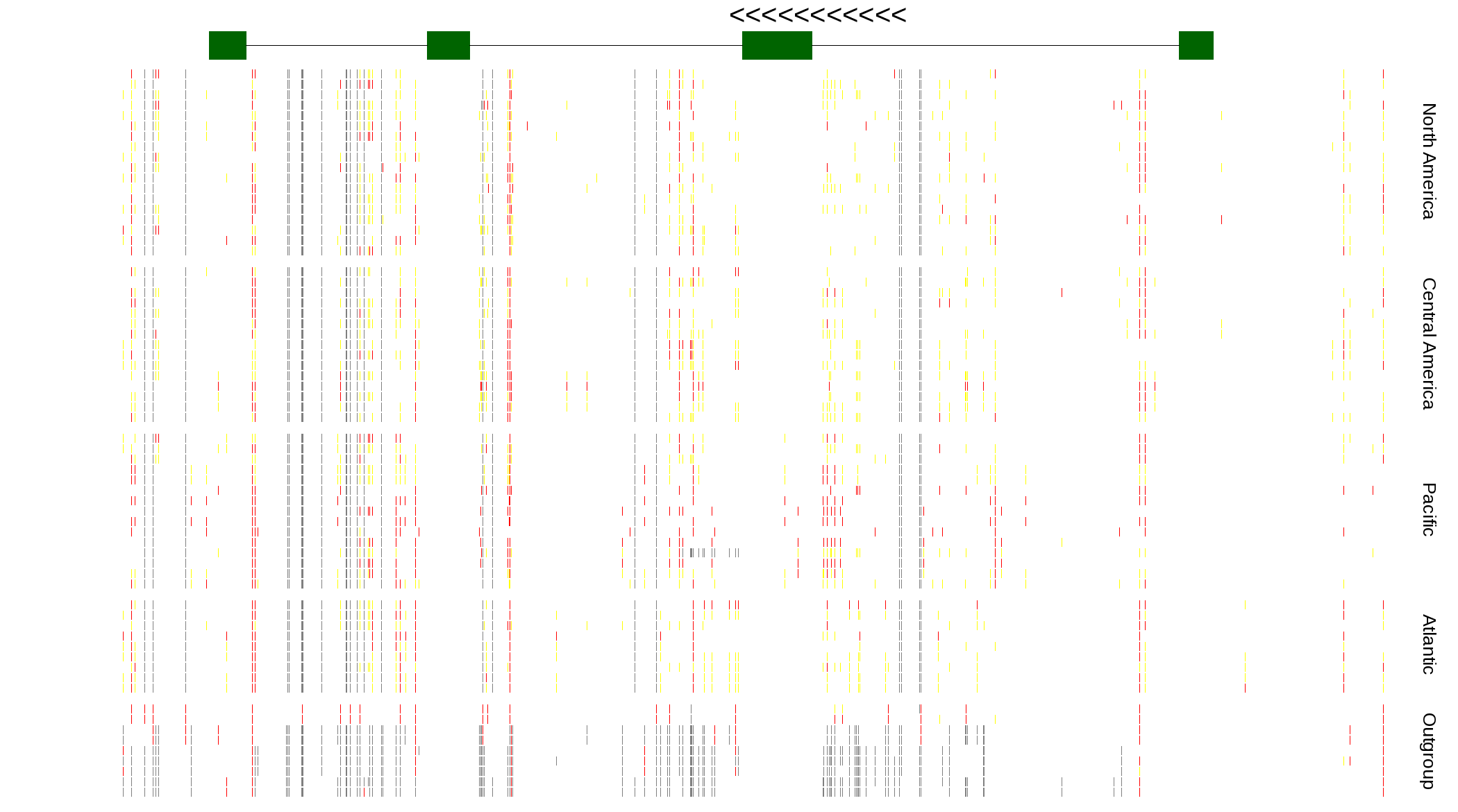
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS200404-TA
ATGACCCCCTTGCCCGCGCCGCCCGTGTTCGACGCCCCCGCAGCCATCCTCACCGTCAGGGACACGGTCAGCTGTAGTTGGATATTTTACAGTAACAGTAGTGGGTCCAACGCGGAGGGCGCCGCCCCGCCGCCGTCTGAACCCGACCCCGACCTGGACTCTATGCACATGATGCTGGACCCGCACCTACGACCGATATCACCAGACCTCACAAACGAAGAGTCCAAACGGATCTTCGACGAACACAAACATCTCGCACAGGAATATCTCAAAATACAAACTGAATTGGCCTACCTCAGCAAACACAAGTCAGAGCTCGAGGAACAAATGGATGGAGAGGAACTGAGACAGAAAAGAGAAATCATACAATTAGAAAATGAGAAGGATTCTTTAATAAAACTGTATTGCACGCTGAAGAAACAACTCACGCGCGCAGATAACGACAGCTGGCTGTTACCGGAGAACGCTCCGCACGAGTGA
>DPOGS200404-PA
MTPLPAPPVFDAPAAILTVRDTVSCSWIFYSNSSGSNAEGAAPPPSEPDPDLDSMHMMLDPHLRPISPDLTNEESKRIFDEHKHLAQEYLKIQTELAYLSKHKSELEEQMDGEELRQKREIIQLENEKDSLIKLYCTLKKQLTRADNDSWLLPENAPHE-